Difference between revisions of "Matlab Examples using MDCS"
| Line 548: | Line 548: | ||
should now be possible. I.e., after resubmission of the job it should run | should now be possible. I.e., after resubmission of the job it should run | ||
properly. | properly. | ||
=== Further troubleshooting === | |||
Say you submitted a job, here referred to as <tt>Job1</tt>, several times | |||
already but it does not finish successfully. Now, if you have no idea how to | |||
troubleshoot further and would like to inform the coordinator of scientific | |||
computing about such problems in order to ask for help, it would be beneficial | |||
and would help in the process of finding the problem if you could provide the | |||
following: | |||
* <tt>Job1.mpiexec.log</tt>-file: If you configured your profile according to the [[MATLAB_Distributing_Computing_Server|mdcs wiki page]] you should find a folder called <tt>Job1</tt> in the <tt>jobData</tt> folder, local to your desktop computer. Within that folder there should be a log-file, in this case called <tt>Job1.mpiexec.log</tt>, e.g., containing information on the nodes that were allocated by the scheduler, the startup/shutdown of smpd daemons on the execution host and the launch of the actual task function you submitted, etc. | |||
* Job-Id: If you happened to have a look at the status of the job via the command <tt>qstat</tt> you also might have recognized the job-id of the job. If you could also provide the job-id, that would even help further. | |||
Revision as of 15:18, 5 July 2013
A few examples for Matlab applications using MDCS (prepared using Matlab version R2011b) are illustrated below.
Example application: 2D random walk
Consider the Matlab .m-file myExample_2DRandWalk.m (listed below), which among other things illustrates the use of sliced variables and independent stremas of random numbers for use with parfor-loops.
This example program generates a number of N independent
2D random walks (a single step has steplength 1 and a
random direction). Each random walk performs tMax steps.
At each step t, the radius of gyration (Rgyr) of walk i
is stored in the array Rgyr_t in the entry Rgyr_t(i,t).
While the whole data is availabe for further postprocessing,
only the average radius of gyration Rgyr_av and the respective
standard error Rgyr_sErr for the time steps 1...tMax are
computed immediately (below it will also be shown how to store the data in an output file on HERO
for further postprocessing).
%% FILE: myExample_2DRandWalk.m
% BRIEF: illustrate sliced variables and independent streams
% of random numbers for use with parfor-loops
%
% DEPENDENCIES:
% singleRandWalk.m - implements single random walk
% averageRgyr.m - computes average radius of gyration
% for time steps 1...tMax
%
% AUTHOR: Oliver Melchert
% DATE: 2013-06-05
%
N = 10000; % number of independent walks
tMax = 100; % number of steps in individual walk
Rgyr_t = zeros(N,tMax); % matrix to hold results: row=radius
% of gyration as fct of time;
% col=independent random walk instances
parfor n=1:N
% create random number stream seeded by the
% current value of n; you can obtain a list
% of all possible random number streams by
% typing RandStream.list in the command window
myStream = RandStream('mt19937ar','Seed',n);
% obtain radius of gyration as fct of time for
% different independent random walks (indepence
% of RWs is ensured by connsidering different
% random number streams for each RW instance)
Rgyr_t(n,:) = singleRandWalk(myStream,tMax);
end
% compute average Rgyr and its standard error for all steps
[Rgyr_av,Rgyr_sErr] = averageRgyr(Rgyr_t);
As liste above, the .m-file depends on the following files:
- singleRandWalk.m, implementing a single random walk, reading:
function [Rgyr_t]=singleRandWalk(randStream,tMax)
% Usage: [Rgyr_t]=singleRandWalk(randStream,tMax)
% Input:
% randStream - random number stream
% tMax - number of steps in random walk
% Output:
% Rgyr_r - array holding the radius of gyration
% for all considered time steps
x=0.;y=0.; % initial walker position
Rgyr_t = zeros(tMax,1);
for t = 1:tMax
% implement random step
phi=2.*pi*rand(randStream);
x = x+cos(phi);
y = y+sin(phi);
% record radius of gyration for current time
Rgyr_t(t)=sqrt(x*x+y*y);
end
end
- averageRgyr.m, which computes the average radius of gyration of the random walks for time steps
1...tMax, reading:
function [avList,stdErrList]=averageRgyr(rawDat)
% Usage: [av]=averageRgyr(rawDat)
% Input:
% rawData - array of size [N,tMax] where N is the
% number of independent random walks and
% tMax is the number of steps taken by an
% individual walk
% Returns:
% av - aveage radius of gyration for the steps
[Lx,Ly]=size(rawDat);
avList = zeros(Ly,1);
stdErrList = zeros(Ly,1);
for i = 1:Ly
[av,var,stdErr] = basicStats(rawDat(:,i));
avList(i) = av;
stdErrList(i) = stdErr;
end
end
function [av,var,stdErr]=basicStats(x)
% usage: [av,var,stdErr]=basicStats(x)
% Input:
% x - list of numbers
% Returns:
% av - average
% var - variance
% stdErr - standard error
av=sum(x)/length(x);
var=sum((x-av).^2)/(length(x)-1);
stdErr=sqrt(var/length(x));
end
For test purposes one might execute the myExample_2DRandWalk.m directly from within a Matlab session on a local Desktop PC.
Specifying file dependencies
So as to sumbit the respective job to the local HPC system one might assemble the following job submission script, called mySubmitScript_v1.m:
sched = findResource('scheduler','Configuration','HERO');
jobRW =...
batch(...
sched,...
'myExample_2DRandWalk',...
'matlabpool',2,...
'FileDependencies',{...
'singleRandWalk.m',...
'averageRgyr.m'...
}...
);
In the above job submission script, all dependent files are listed as FileDependencies. I.e.,
the .m-files specified therein are copied from your local desktop PC to the HPC system at run time.
Now, from within a Matlab session I navigate to the Folder where the above .m-files are located in and call the job submission script, i.e.:
>> cd MATLAB/R2011b/example/myExamples_matlab/RandWalk/ >> mySubmitScript_v1 runtime = 24:0:0 (default) memory = 1500M (default) diskspace = 50G (default)
before the job is actually submitted, I need to specify my user ID and password,
of course. Once the job is successfully submitted, I can check the state of the
job via typing jobRW.state. However, if you want to get some more information
on the status of your job, you might want to try to log-in on the HPC system and to
simply type the command qstat on the commandline. This will yield several
details related to your job which you might process further to see on which execution nodes your
job runs, why it won't start directly etc. Note that MATLAB provides only a wrapper for
the qstat command which in some cases result in a misleading output. E.g.,
if, for some reason, your job changes to the error-state it might be that MATLAB erroneously
reports it to be in the finished-state.
Once the job (really) has finished, i.e.,
>> jobRW.state ans = finished
I might go on and load the results to my desktop computer, giving
>> res=load(jobRW);
>> res
res =
N: 10000
Rgyr_av: [100x1 double]
Rgyr_sErr: [100x1 double]
Rgyr_t: [10000x100 double]
ans: 'finished'
res: [1x1 struct]
tMax: 100
However, note that there are several drawbacks related to the usage of FileDependencies: E.g.,
- each worker gets an own copy of the respective .m-files when the job starts (in particular, workers that participate in the computing process do not share a set of .m-files in a common location),
- the respective .m-files are not available on the HPC system once the job has finished,
- comparatively large input files need to be copied to the HPC system over and over again, if several computations on the same set of input data are performed.
In many cases a different procedure, based on specifying PathDependencies, outlined below in detail, might be recommendend.
Specifying path dependencies
Basically there are two ways to specify path dependencies. You might either specify them in your job submission script or directly in your main MATLAB .m-file. Below, both approaches are illustrated.
Modifying the job submission script
The idea underlying the specification of path dependencies is that there might be MATLAB modules (or
sets of data) you want to routinely use over and over again. Then, having these modules
available on your local desktop computer and using file dependencies to copy them
to the HPC system at run time results in time and memory consuming, unnecessary operations.
As a remedy you might adopt the following two-step procedure:
- copy the respective modules to the HPC system
- upon submitting the job from your local desktop pc, indicate by means of the key word
pathDependencieswhere (on HERO) the respective data files can be found.
This eliminates the need to copy the respective files using the fileDependencies statement.
An example of how to accomplish this for the random walk example above is given below.
Just for arguments, say, the content of the two module files singleRandWalk.m and averageRgyr.m
will not be changed in the near future and you want to use both files on a regular basis when you submit jobs to the HPC cluster.
Hence, it would make sense to copy them to the HPC system and to specify within your job submission script where they can
be found for use by any execution host. Following the above two steps you might proceed as follows:
1. create a folder where you will copy both files to. To facilitate intuition and to make this as explicit as possible, I created the folder
/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/random_walk
and copied both files there, see
alxo9476@hero01:~/SIM/MATLAB/R2011b/my_modules/random_walk$ ls averageRgyr.m singleRandWalk.m
2. Now, it is no more necessary to specify both files as file dependencies as in the example above. Instead, in your job submission file (here called mySumbitScript_v2.m) you might now specify a path depenency (which relates to your filesystem on the HPC system) as follows:
sched = findResource('scheduler','Configuration','HERO');
jobRW =...
batch(...
sched,...
'myExample_2DRandWalk',...
'matlabpool',2,...
'PathDependencies',{'/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/random_walk'}...
);
This has the benefit that the files will not be copied to the HPC system at run time and that there is only a single copy of those files on the HPC system, which can be used by all execution hosts (so no multiple copies of the same files necessarry as with the use of file dependencies).
Again, from within a Matlab session navigate to the folder where the job submission file and the main file myExample_2DRandWalk.m are located in and call the job submission script. For me, this reads:
>> cd MATLAB/R2011b/example/myExamples_matlab/RandWalk/ >> mySubmitScript_v2 runtime = 24:0:0 (default) memory = 1500M (default) diskspace = 50G (default)
again, before the job is actually submitted, I need to specify my user ID and password (since I started a new MATLAB session in between). Once the job has finished I might go on and load the results to my desktop computer, giving
>> res=load(jobRW);
>> res
res =
N: 10000
Rgyr_av: [100x1 double]
Rgyr_sErr: [100x1 double]
Rgyr_t: [10000x100 double]
ans: 'finished'
res: [1x1 struct]
tMax: 100
Modifying the main m-file
As an alternative to the above procedure, you might add the folder
/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/
to your local MATLAB path by adding the single line
addpath(genpath('/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/'));
to the very beginning of the file myExample_2DRandWalk.m. For completeness, note
that this adds the above folder my_modules and all its subfolders
to your MATLAB path. Consequently, all the m-files contained therein will be
available to the execution nodes which contribute to a MATLAB session.
In this case, a proper job submission script (mySubmitScript_v3.m) reads simply:
sched = findResource('scheduler','Configuration','HERO');
jobRW =...
batch(...
sched,...
'myExample_2DRandWalk',...
'matlabpool',2...
);
As a personal comment, note that, from a point of view of submitting a job to the HPC System, I would always prefer the explicit way of stating path dependencies in the job submission file over the implicit way of indirectly implying them using a modification of the main m-file. The latter choice seems much more vulnerable to later changes!
Storing data on HERO
Consider a situation where your application produces lots of data you want to store for further postprocessing. Often, in particular when you produce lots of data, you also don't want the data to be copied back to your desktop computer, immediately. Instead you might want to store the data on HERO and perhaps mount your HPC-homedirectory later to relocate the data (or the like). Note that, as a default HERO user, you have 110GB of disk space available under your home directory. Below it is illustrated, by means of the random walk example introduced earlier, how to store output data on the HPC system.
The only thing you need to do is to specify a path within your main m-file under which the data should be stored. Therefore you first have to create the corresponding sequence of folders if they do not exist already. To facilitate intuition: in my case I decided to store the data under the path
/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_data/random_walk/
I created the folder my_data/rand_walk/ for that purpose. Right now, the folder is empty.
In the main file (here called myExample_2DRandWalk_saveData.m) I implemented the following changes:
N = 10000; % number of independent walks
tMax = 100; % number of steps in individual walk
Rgyr_t = zeros(N,tMax); % matrix to hold results: row=radius
% of gyration as fct of time;
% col=independent random walk instances
% absolute path to file on HERO where data will be saved
outFileName=sprintf('/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_data/random_walk/rw2d_N%d_t%d.dat',N,tMax);
parfor n=1:N
% create random number stream seeded by the
% current value of n; you can obtain a list
% of all possible random number streams by
% typing RandStream.list in the command window
myStream = RandStream('mt19937ar','Seed',n);
% obtain radius of gyration as fct of time for
% different independent random walks (indepence
% of RWs is ensured by connsidering different
% random number streams for each RW instance)
Rgyr_t(n,:) = singleRandWalk(myStream,tMax);
end
% compute average Rgyr and its standard error for all steps
[Rgyr_av,Rgyr_sErr] = averageRgyr(Rgyr_t);
% write data to output file on HERO
saveData_Rgyr(outFileName,Rgyr_av, Rgyr_sErr);
Note that the outFileName is specified directly in the main m-file (there are also more elegant ways to accomplish this, however, for the moment this will do!) and the new function, termed saveData_Rgyr is called. The latter function will just write out
some statistical summary measures related to the gyration radii of the 2D random walks. For completeness, it reads:
function saveData(fileName,Rgyr_av,Rgyr_sErr)
% Usage: saveData(fileName,myData)
% Input:
% fileName - name of output file
% myData - data to be saved in file
% Returns: nothing
outFile = fopen(fileName,'w');
fprintf(outFile,'# t Rgyr_av Rgyr_sErr \n');
for i = 1:length(Rgyr_av)
fprintf(outFile,'%d %f %f\n',i,Rgyr_av(i),Rgyr_sErr(i));
end
fclose(outFile);
end
Now, say, my opinion on the proper output format is not settled yet and I consider to experiment with different kinds of output formatting styles, for that matter. Then it is completely fine to specify some of the dependent files as path dependencies (namely those that are unlikely to change soon) and others as file dependencies (namely those which are under development). Here, joining in the specification of path dependencies within a job submission script, a proper submission script (here called mySubmitScript_v4.m) might read:
sched = findResource('scheduler','Configuration','HERO');
jobRW =...
batch(...
sched,...
'myExample_2DRandWalk_saveData',...
'matlabpool',2,...
'FileDependencies',{'saveData_Rgyr.m'},...
'PathDependencies',{'/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/random_walk'}...
);
Again, starting a MATLAB session, changing to the directory where the myExample_2DRandWalk_saveData.m and saveData_Rgyr.m files are located in, launching the submission script mySubmitScript_v5.m and waiting for the job to finish, i.e.
>> cd MATLAB/R2011b/example/myExamples_matlab/RandWalk/ >> mySubmitScript_v4 runtime = 24:0:0 (default) memory = 1500M (default) diskspace = 50G (default) >> jobRW.state ans = finished
Once the job is done, the output file rw2d_N10000_t100.dat (as specified in the main script myExample_2DRandWalk_saveData.m)
is created in the folder under the path
/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_data/random_walk/
which contains the output data in the format as implemented by the function saveData_Rgyr.m, i.e.
alxo9476@hero01:~/SIM/MATLAB/R2011b/my_data/random_walk$ head rw2d_N10000_t100.dat # t Rgyr_av Rgyr_sErr 1 1.000000 0.000000 2 1.273834 0.006120 3 1.572438 0.007235 4 1.795441 0.008645 5 1.998900 0.009714 6 2.168326 0.010867 7 2.355482 0.011766 8 2.519637 0.012721 9 2.683048 0.013456
Again, after the job has finished, the respective file remains on HERO and is available for further postprocessing. It is not automatically copied to your local desktop computer.
Postprocessing of the simulated data
Just for completeness a brief analysis and display of the 2D random walk data is presenter here.
Once the job has finished and the resulting data is loaded to the desktop
computer via res=load(jobRW); its possible to postprocess and display
some of the results via the m-file finalPlot.m which reads:
function finalPlot(res)
% Usage: finalPlot(res)
% Input:
% res - simulation results for 2D rand walks
% Returns: (none)
%
subplot(1,2,1), plotHelper(res);
subplot(1,2,2), histHelper(res);
end
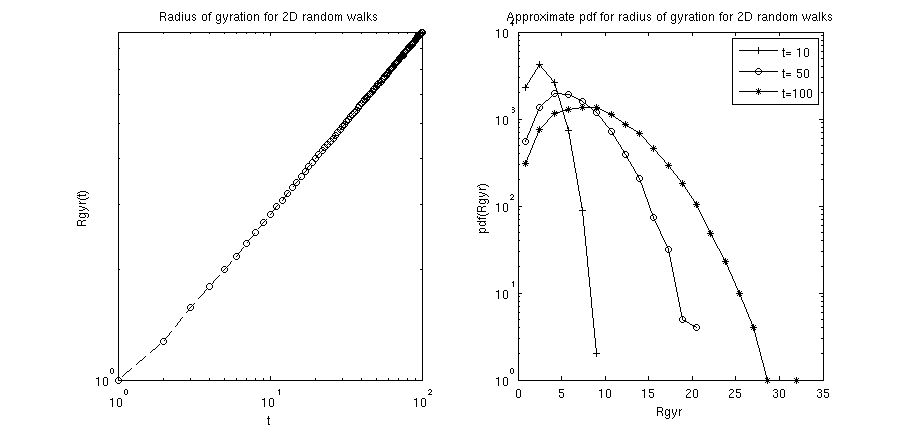
function plotHelper(res)
% Usage: plotHelper(res)
%
% implements plot of the gyration radius as
% function of the walk length
%
% Input:
% res - simulation results for 2D rand walks
% Returns: (none)
%
xRange=1:1:100;
loglog(xRange,res.Rgyr_av,'--ko');
title('Radius of gyration for 2D random walks');
xlabel('t');
ylabel('Rgyr(t)');
end
function histHelper(res)
% Usage: histHelper(res)
%
% implements histogram of the pdf for gyration
% radii at chosen times steps
% of the walk length
%
% Input:
% res - simulation results for 2D rand walks
% Returns: (none)
%
set(0,'DefaultAxesColorOrder',[0 0 0],...
'DefaultAxesLineStyleOrder','-+|-o|-*');
tList=[10,50,100];
[events,key]=hist(res.Rgyr_t(:,tList),20);
semilogy(key,events);
title('Approximate pdf for radius of gyration for 2D random walks');
xlabel('Rgyr');
ylabel('pdf(Rgyr)');
key = legend('t= 10','t= 50','t=100');
end
Therein, the main function calls two subfunctions:
plotHelper(res): implements the gyration radius of the 2D random walks as a function of the number of steps in the walk (i.e. the elapsed time since the walk started). The radius of gyration of such an unhindered 2D random walks generally scales (for not too short times ).histHelper(res): implements histograms of the probability density function (pdf) for the gyration radii at specified time steps. In the above example the time steps and where chosen. For not too short times, one can expect that the pdf is properly characterized by the Rayleigh distribution .
Calling the m-file should yield a figure similar to the following:
Other Topics
Permision denied upon attempted passphraseless login on remote host
Consider the following situation: you submit a Job, employing an overall number of three workers, from your local client to the remote HPC system and SGE places the job in an appropriate queue (following your resource specifications). Just for arguments: say, the job got the name Job118. After a while the job changes from its state qw to r and to dr soon after, so that the job is not executed properly.
In your /MATLAB/R2011b/jobData folder on the HPC system (during the runtime of the job), or on your local client (if the job has already finished and the respective files are transfered to your client) there should be a folder called Job118, wherein a log-file can be found. For the job Job118, this log-file is called Job118.mpiexec.log and it might contain some information on why the job is not executed properly. Now, say the content of Job118.mpiexec.log reads something similar to
Starting SMPD on mpcs005.mpinet.cluster mpcs103.mpinet.cluster mpcs111.mpinet.cluster ... /cm/shared/apps/sge/6.2u5p2/mpi/mdcs/start_mdcs -n mpcs005.mpinet.cluster "/cm/shared/apps/matlab/r2011b/bin/mw_smpd" 25327 /cm/shared/apps/sge/6.2u5p2/mpi/mdcs/start_mdcs -n mpcs103.mpinet.cluster "/cm/shared/apps/matlab/r2011b/bin/mw_smpd" 25327 /cm/shared/apps/sge/6.2u5p2/mpi/mdcs/start_mdcs -n mpcs111.mpinet.cluster "/cm/shared/apps/matlab/r2011b/bin/mw_smpd" 25327 Permission denied, please try again. Permission denied, please try again. Check for smpd daemons (1 of 10) Permission denied (publickey,gssapi-with-mic,password). Warning: Permanently added 'mpcs005.mpinet.cluster,10.142.3.5' (RSA) to the list of known hosts. Warning: Permanently added 'mpcs111.mpinet.cluster,10.142.3.111' (RSA) to the list of known hosts. Missing smpd on mpcs005.mpinet.cluster Missing smpd on mpcs103.mpinet.cluster Missing smpd on mpcs111.mpinet.cluster Permission denied, please try again. Permission denied, please try again. Permission denied, please try again. Permission denied, please try again. Permission denied (publickey,gssapi-with-mic,password). Permission denied (publickey,gssapi-with-mic,password). Check for smpd daemons (2 of 10) Missing smpd on mpcs005.mpinet.cluster Missing smpd on mpcs103.mpinet.cluster Missing smpd on mpcs111.mpinet.cluster
This might indicate that the passphraseless login to the execution nodes, here the nodes mpcs005.mpinet.cluster mpcs103.mpinet.cluster mpcs111.mpinet.cluster on which the workers should be started, has failed.
Albeit there are many reasons that might cause such a behavior, one that seems likely is the following: you probably changed the rights of your home directory in a way that also the group and/or others have the right to write to your home directory. To make this as explicit as possible, I change my rights accordingly to give an example: so, in my homedirectory on HERO I type
ls -lah ./
to find
drwxrwxrwx 14 alxo9476 ifp 937 Jul 2 12:19 .
considering the ten leftmost letters this is bad. The first d tells that we are looking at a directory. The following three letters rwx tell that the owner of the directory has the rights to read, write, and execute. The following three letters signify that the group has the right to read, write, and execute and the last three letters indicate that all others have the right to read, write, and execute files. Note that for a successful passphraseless login on a remote host, it must be made sure that only the owner of the remote users home directory (which in this case are you, yourself!), has the right to write.
As a remedy try the following: To resore the usual rights in my homedirectory on HERO I type
chmod 755 ./
and now find
drwxr-xr-x 14 alxo9476 ifp 937 Jul 2 12:19 .
which shows that the writing rights for the unix group I belong to (in my case ifP) and all others are removed. Accordingly, a passphraseless login to the hosts specified in the Hostfile should now be possible. I.e., after resubmission of the job it should run properly.
Further troubleshooting
Say you submitted a job, here referred to as Job1, several times already but it does not finish successfully. Now, if you have no idea how to troubleshoot further and would like to inform the coordinator of scientific computing about such problems in order to ask for help, it would be beneficial and would help in the process of finding the problem if you could provide the following:
- Job1.mpiexec.log-file: If you configured your profile according to the mdcs wiki page you should find a folder called Job1 in the jobData folder, local to your desktop computer. Within that folder there should be a log-file, in this case called Job1.mpiexec.log, e.g., containing information on the nodes that were allocated by the scheduler, the startup/shutdown of smpd daemons on the execution host and the launch of the actual task function you submitted, etc.
- Job-Id: If you happened to have a look at the status of the job via the command qstat you also might have recognized the job-id of the job. If you could also provide the job-id, that would even help further.