Difference between revisions of "Gaussian 2016"
| (138 intermediate revisions by 4 users not shown) | |||
| Line 5: | Line 5: | ||
== Installed version == | == Installed version == | ||
These currently installed versions are available... | |||
... on envirnoment ''hpc-uniol-env'': | |||
'''gaussian/g09.b01''' | |||
'''gaussian/g09.d01''' | |||
'''gaussian/g16.a03''' | |||
'''gaussian/g16.C01''' | |||
... on environment ''hpc-env/6.4'': | |||
'''Gaussian/g16.a03-AVX2''' | |||
'''gaussian/g16.a03''' | |||
'''gaussian/g16.c01''' | |||
We recommend using the environment <tt>hpc-env/6.4</tt> as in general more recent software versions are available. If you also want to use GaussView you may have to as the required graphics libraries are not available otherwise. | |||
== Available abilities == | == Available abilities == | ||
According to the official homepage, Gaussian 09 Rev. D01 has the following abilities: | |||
*Molecular mechanics | *Molecular mechanics | ||
| Line 28: | Line 43: | ||
*Quantum chemistry composite methods (CBS-QB3, CBS-4, CBS-Q, CBS-Q/APNO, G1, G2, G3, W1 high-accuracy methods) | *Quantum chemistry composite methods (CBS-QB3, CBS-4, CBS-Q, CBS-Q/APNO, G1, G2, G3, W1 high-accuracy methods) | ||
== Using Gaussian 09. | '''Note:''' Rev. D.01 has a bug that can cause Gaussian jobs to fail during geometry optimization. See below (or click [https://wiki.hpcuser.uni-oldenburg.de/index.php?title=Gaussian_2016#Current_Issues_with_Gaussian_09_Rev._D.01 here]) for details and possible work-arounds. | ||
== Using Gaussian on the HPC cluster == | |||
If you want to find out more about Gaussian on the HPC cluster, you can use the command | |||
module spider gaussian | |||
which will give you an output looking like this: | |||
----- /cm/shared/uniol/modulefiles/chem ----- | |||
... gaussian/g09.b01 ... | |||
To load a specific version of Gaussian use the full name of the module, e.g. to load Gaussian 09 Rev. D.01 (this is also the default): | |||
[abcd1234@hpcl001 ~]$ module load gaussian/g09.d01 | |||
[abcd1234@hpcl001 ~]$ module list | |||
Currently loaded modules: ... gaussian/g09.d01 ... | |||
=== Single-node (multi-threaded) jobs === | |||
'''Example:''' The following job script <tt>run_gaussian.job</tt> defines a single-node job named ''g09test'' on the partition ''carl.p'', using ''8 cpus'' (cores), a total of ''6 GB of RAM'' and ''100G of local storage (<tt>$TMPDIR</tt>)'' for a maximum run time of ''2 hours'': | |||
#!/bin/bash | |||
#SBATCH --job-name=g09test # job name | |||
#SBATCH --partition=carl.p # partition | |||
#SBATCH --time=0-2:00:00 # wallclock time d-hh:mm:ss | |||
#SBATCH --ntasks=1 # only one task for G09 | |||
#SBATCH --cpus-per-task=8 # multiple cpus per task | |||
#SBATCH --mem=6gb # total memory per node | |||
#SBATCH --gres=tmpdir:100G # reserve 100G on local /scratch | |||
INPUTFILE=dimer | |||
# load module for Gaussian version to be used (here g09.d01) | |||
module load gaussian/g09.d01 | |||
# call g09run which now takes up to two arguments | |||
# <inputfile>: name of the inputfile (required) | |||
# [setnproc]: control whether %NProcShared= is set to match | |||
# the requested number of CPUs above (optional) | |||
# 0 - do not change the input file (default) | |||
# 1 - modify %NProcShared=N if N is larger than | |||
# number of CPUs per task requested | |||
# 2 - modify %NProcShared=N if N is not equal | |||
# to the number of CPUs per task requested | |||
g09run $INPUTFILE 2 | |||
The example job script and the used input file can be downloaded [[Media:Gaussian_testjob+example.zip | here]] (.zip-file). | |||
To submit this job, you have to use the following command: | |||
[abcd1234@hpcl001 ~]$ sbatch run_gaussian.job | |||
Submitted batch job 54321 | |||
The job will start running as soon as the requested ressources can be allocated. The output of the job (including a checkpoint file in <tt>$HOME/g09/CHK</tt>) will be produced in the normal directories. | |||
'''Note''': The above procedure and the script are the same for Gaussian 16, except that <tt>g09run</tt> has to be replaced by <tt>g16run</tt>. Please read the [http://gaussian.com/relnotes/ release notes] to find out what has changed in Gaussian 16. | |||
'''Explanations''' for the job script: | |||
The expected run time of a job is requested with the <tt>--time</tt> option e.g. in the format <tt>d-hh:mm:ss</tt> (you can also omit the <tt>:ss</tt> part). The default is 2 hours if nothing else is specified in the job script or at job submission. | |||
On the old cluster, you had to request a parallel environment by adding a line '<tt> #$ -pe smp 12</tt>' to your job script for a single node job. SLURM does not have parallel environments but you can request CPUs (cores) per task to the same effect. The following settings in the above job script serve as an example: | |||
#SBATCH --partition=carl.p | |||
#SBATCH --ntasks=1 | |||
#SBATCH --cpus-per-task=8 | |||
The maximum amount of "cpus per task" varies for the different partitions (node types) are: | |||
*'''carl.p''' = max. 24 cpus per task | |||
*'''mpcl.p''' = max. 24 cpus per task | |||
*'''mpcs.p''' = max. 24 cpus per task | |||
*'''mpcb.p''' = max. 16 cpus per task | |||
*'''mpcp.p''' = max. 40 cpus per task | |||
So, if you want to run a single node job on a MPC-BIG node using all available cores, the lines above should be changed to: | |||
#SBATCH --partition=mpcb.p | |||
#SBATCH --ntasks=1 | |||
#SBATCH --cpus-per-task=16 | |||
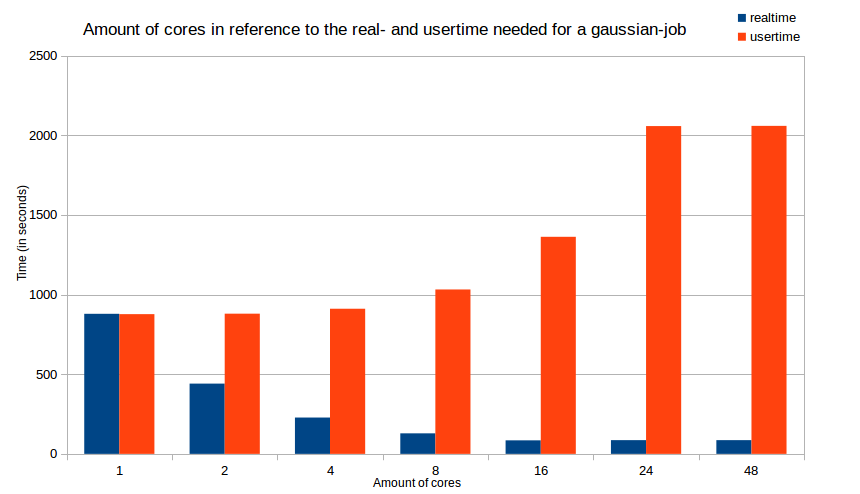
'''Note''': The numbers mentioned above are the maximum amount of CPUs for each node. You can use them but its not always profitable to do so. It is always a good idea to make some tests to find the optimal number of cores. An example is shown in this [[Gaussian 2016#Optimizing the runtime of your jobs | diagramm]] which shows the real (wall clock) and user (CPU) time as a function of number of CPUs (cores). | |||
In the example, memory is requested per node with the <tt>--mem</tt>-option. This is more practical for Gaussian jobs than requesting memory per core which is also possible with the <tt>--mem-per-cpu</tt>-option. | |||
Every node has additional local storage which can be used while your job is running. The following line in the example job script | |||
#SBATCH --gres:tmpdir:100G | |||
requests the use of 100GB of local storage for your job. The path for this space is $TMPDIR as always. Further informations can be found [[File system and Data Management#Scratch space / TempDir | here]]. | |||
== | === Memory Considerations === | ||
With the line | |||
# | #SBATCH --mem 6Gb | ||
the total amount of memory reserved for the job is 6GB (alternative, use <tt>--mem-per-cpu</tt> to allocate memory per CPU core requested). In the partition <tt>carl.p</tt> you can reserve up to 117GB per node. You can find out the limits for other partitions using | |||
scontrol show partition <partname.p> | |||
</ | and look for the parameter <tt>MaxMemPerNode</tt> (the value is in MB). Note, that nodes can be shared between different jobs and the smaller your memory limit is, so more likely your job can run on shared node. '''Note''': if you are not requesting memory explicitly, your job will receive a default amount per requester CPU core. In many cases this may be sufficient and if you are using all cores on a node you also get all available memory. | ||
The | The Gaussian input files also contain, e.g., the following line in the link 0 section: | ||
%Mem=21000MB | |||
%Mem=21000MB | to indicate to Gaussian the available memory. | ||
'''Important''': Memory management is critical for the performance of Gaussian jobs. Which parameter values are optimal is highly dependent on the type of the calculation, the system size, and other factors. Therefore, optimizing your Gaussian job with respect to memory allocation almost always requires (besides experience) some trial and error. The following general remarks may be useful: | '''Important''': Memory management is critical for the performance of Gaussian jobs. Which parameter values are optimal is highly dependent on the type of the calculation, the system size, and other factors. Therefore, optimizing your Gaussian job with respect to memory allocation almost always requires (besides experience) some trial and error. The following general remarks may be useful: | ||
== Linda | In the above example, we have told Gaussian to use almost 21GB (divide MB by 1024) for the job, which means we need to request at least that much for job as well. In fact, since the G09 executables are rather large and have to be resident in memory, there should be some margin (at least 1GB, better 10%, therefore for the above link 0 command we would use <tt>--mem 23Gb</tt>). This is usually a good choice for DFT calculations. | ||
For MP2 calculations, on the other hand, Gaussian requests about twice the amount of memory specified by the <tt>Mem=...</tt> directive. If it exceeds the memory reserved for the job, the scheduler will terminate the job. Therefore, for a MP2 calculation the link 0 line should read: | |||
%Mem=11000MB | |||
if 23GB have been requested for the job. | |||
=== Linda Jobs === | |||
Gaussian multi-node jobs (Linda-jobs) can be submitted by simply changing the "--ntasks"-value to something > 1, e.g. | |||
#SBATCH --ntasks=2 | |||
Everything else will be handled by the <tt>g09run</tt>-script, you must no longer add "'''%LindaWorkers='''" to your inputfile! | |||
'''Note:''' Linda is only available for Gaussian 09. Gaussian 16 is limited to single node jobs (up to 40 cores in the partition <tt>mpcp.p</tt>). | |||
== Letting g09run set the number of processes == | |||
If you have used Gaussian on the old cluster, you will already know what the command "g09run" in your jobscript does: its runs Gaussian with a given input file. This remains unchanged on the new cluster. On the new cluster CARL a feature has been added to <tt>g09run</tt>. The new feature is simple, but very effective for saving resources that would otherwise be "wasted". We have implemented the possibility to add another option beside naming the input file to "g09run" | |||
'''setnproc''' has three different options: | |||
*0: do not change the input file (default) | |||
*1: modify %NProcShared=N if N is larger than number of CPUs per task requested | |||
*2: modify %NProcShared=N if N is not equal to the number of CPUs per task requested | |||
The full syntax of the "g09run"-command looks like this: | |||
g09run <INPUTFILE> [SETNPROC] | |||
where the first argument is mandatory and second optional (i.e. if you do not want to use the new feature you can ignore it). | |||
'''Example''': | |||
Using | |||
g09run my_input_file 1 | |||
in your job script will cause <tt>g09run</tt> to check whether <tt>%NProcShared</tt> is too large for the requested number of CPUs and modify it if needed. | |||
== Optimizing the runtime of your jobs == | |||
Optimizing the runtime of your jobs will not only safe your time, it will also safe resources (cores, memory, time etc.) for everyone else using the cluster. Therefore you should determine the best amount of cores for your particular job. The following diagram will show the time difference (splitted into real (wall clock) time and user (CPU) time) in terms of the amount of used cores/CPUs. The job we used to gather these times is the example job mentioned above. | |||
[[Image:Dia cores time.png | Cores in reference to real and user time]] | |||
(Every test job was done with 6 GB of RAM, except the one with 48 cores which was done with 12 GB of RAM) | |||
As you see in the diagram, increasing the amount of cores will reduce the time your job needs to finish (= ''real time''). However, if you reach a certain amount of cores (16 cores in this example), adding cores will not significantly decrease the real time, it will just increase the time your job is processed by the cpus (= ''user time''). The user time peaks at about 16 cores too, so adding even more cores is not beneficial in any ways. | |||
With '''Gaussian 16''' a Link 0-option has been added that allows you to bind the processes to individual CPU-cores with the aim to improve performance. Binding processes to a core has the advantage that memory access from that process may be more efficient. This can be achieved by replacing the Link 0-line | |||
%nprocshared=24 | |||
with the new option | |||
%CPU=0-23 | |||
Now Gaussian will at runtime bind each parallel process to a single core rather than letting the OS decide which process to run on which core. In this example, a complete node with 24 cores must be requested. | |||
If in the job script you are requesting less cores than available per node, e.g. | |||
#SBATCH --ntasks=1 # only one task for G16 | |||
#SBATCH --cpus-per-task=8 # multiple cpus per task | |||
only 8 cores in this example, than the problem is is that you cannot know in advance which cores will be assigned to you. However, writing | |||
%CPU=0-7 | |||
< | will tell Gaussian to use only the cores with the core IDs 0 to 7. If you get assigned cores with IDs 2,4,5-8,10-11 the above Link 0-option will cause the following error | ||
< | Error: MP_BLIST has an invalid value | ||
indicating that Gaussian tried to bind a process to a core without having the permission to do so. You Link 0-option needs to be | |||
%CPU=2,4,5-8,10-11 | |||
which obviously requires you to modify your input file at runtime. Here is an example how to: | |||
INPUTFILE=dimer | |||
# update inputfile for CPUs | |||
CPUS=$(taskset -cp $$ | awk -F':' '{print $2}') | |||
sed -i -e "/^.*%cpu/I s/=.*$/=$CPUS/" $INPUTFILE | |||
This assumes your inputfile already has a line starting with <tt>%CPU=</tt> and uses the taskset command to find out, which cores are available for your job. | |||
== Current | == Current issues with Gaussian 09 Rev. D.01 == | ||
Currently Rev. D.01 has a bug which can cause geometry optimizations to fail. If this happens, the following error message will appear in your log- or out-file: | Currently Rev. D.01 has a bug which can cause geometry optimizations to fail. If this happens, the following error message will appear in your log- or out-file: | ||
| Line 123: | Line 219: | ||
Error termination in NtrErr: | Error termination in NtrErr: | ||
NtrErr Called from FileIO. | NtrErr Called from FileIO. | ||
The explanation from Gaussian's technical support: | '''The explanation from Gaussian's technical support:''' | ||
<blockquote> | <blockquote> | ||
This problem appears in cases where one ends up with different orthonormal subsets of basis functions at different geometries. The "Operation on file out of range" message appears right after the program tries to do an interpolation of two SCF solutions when generating the initial orbital guess for the current geometry optimization point. The goal here is to generate an improved guess for the current point but it failed. The interpolation of the previous two SCF solutions to generate the new initial guess was a new feature introduced in G09 rev. D.01. The reason why this failed in this particular case is because the total number of independent basis functions is different between the two sets of orbitals. We will have this bug fixed in a future Gaussian release, so the guess interpolation works even if the number of independent basis functions is different. | This problem appears in cases where one ends up with different orthonormal subsets of basis functions at different geometries. The "Operation on file out of range" message appears right after the program tries to do an interpolation of two SCF solutions when generating the initial orbital guess for the current geometry optimization point. The goal here is to generate an improved guess for the current point but it failed. The interpolation of the previous two SCF solutions to generate the new initial guess was a new feature introduced in G09 rev. D.01. The reason why this failed in this particular case is because the total number of independent basis functions is different between the two sets of orbitals. We will have this bug fixed in a future Gaussian release, so the guess interpolation works even if the number of independent basis functions is different. | ||
</blockquote> | </blockquote> | ||
There a number of suggestions from the technical support on how to work around this problem: | '''There a number of suggestions from the technical support on how to work around this problem:''' | ||
<blockquote> | <blockquote> | ||
A) Use “Guess=Always” to turn off this guess interpolation feature. | '''A)''' Use “Guess=Always” to turn off this guess interpolation feature. | ||
Option "A" would work in many cases, although it may not be a viable alternative in cases where the desired SCF solution is difficult to get from the default guess and one has to prepare a special initial guess. You may try this for your case. | Option "A" would work in many cases, although it may not be a viable alternative in cases where the desired SCF solution is difficult to get from the default guess and one has to prepare a special initial guess. You may try this for your case. | ||
B) Just start a new geometry optimization from that final point reading the geometry from the checkpoint file. | '''B)''' Just start a new geometry optimization from that final point reading the geometry from the checkpoint file. | ||
Option "B" should work just fine although you may run into the same issue again if, after a few geometry optimization steps, one ends up again in the scenario of having two geometries with two different numbers of basis functions. | Option "B" should work just fine although you may run into the same issue again if, after a few geometry optimization steps, one ends up again in the scenario of having two geometries with two different numbers of basis functions. | ||
C) Lower the threshold for selecting the linearly independent set by one order of magnitude, which may result in keeping all functions. The aforementioned threshold is controlled by "IOp(3/59=N)" which sets the threshold to 1.0D-N (N=6 is the default). Note that because an IOp is being used, one would need to run the optimization and frequency calculations separately and not as a compound job ("Opt Freq"), because IOps are not passed down to compound jobs. You may also want to use “Integral=(Acc2E=11)” or “Integral=(Acc2E=12)” if you lower this threshold as the calculations may not be as numerically robust as with the default thresholds. | '''C)''' Lower the threshold for selecting the linearly independent set by one order of magnitude, which may result in keeping all functions. The aforementioned threshold is controlled by "IOp(3/59=N)" which sets the threshold to 1.0D-N (N=6 is the default). Note that because an IOp is being used, one would need to run the optimization and frequency calculations separately and not as a compound job ("Opt Freq"), because IOps are not passed down to compound jobs. You may also want to use “Integral=(Acc2E=11)” or “Integral=(Acc2E=12)” if you lower this threshold as the calculations may not be as numerically robust as with the default thresholds. | ||
Option "C" may work well in many cases where there is only one (or very few) eigenvalue of the overlap matrix that is near the threshold for linear dependencies, so it may just work fine to use "IOp(3/59=7)", which will be keeping all the functions. Because of this situation, and because of potential convergence issues derived from including functions that are nearly linearly dependent, I strongly recommend using a better integral accuracy than the default, for example "Integral=(Acc2E=12)", which is two orders of magnitude better than default. | Option "C" may work well in many cases where there is only one (or very few) eigenvalue of the overlap matrix that is near the threshold for linear dependencies, so it may just work fine to use "IOp(3/59=7)", which will be keeping all the functions. Because of this situation, and because of potential convergence issues derived from including functions that are nearly linearly dependent, I strongly recommend using a better integral accuracy than the default, for example "Integral=(Acc2E=12)", which is two orders of magnitude better than default. | ||
D) Use fewer diffuse functions or a better balanced basis set, so there aren’t linear dependencies with the default threshold and thus no functions are dropped. | '''D)''' Use fewer diffuse functions or a better balanced basis set, so there aren’t linear dependencies with the default threshold and thus no functions are dropped. | ||
Option "D" is good since it would avoid issues with linear dependencies altogether, although it has the disadvantage that you would not be able to reproduce other results with the basis set that you are using. | Option "D" is good since it would avoid issues with linear dependencies altogether, although it has the disadvantage that you would not be able to reproduce other results with the basis set that you are using. | ||
</blockquote> | </blockquote> | ||
== Changes from G09 to G16 == | |||
Here is an overview of changes between G09 and G16 in general and regarding the use of the cluster: | |||
# Changes of the latest version of Gaussian can always be found in the [http://gaussian.com/relnotes/ release notes]. | |||
# Gaussian 16 can be used on the cluster by loading the corresponding module and using <tt>g16run</tt> within the job script. | |||
# Link 0 (%-lines) inputs can now be specified in different ways (see [http://gaussian.com/relnotes/?tabid=5 Link 0 Equivs.]), which in principle should work on the cluster as well (with the exception of command-line arguments if you use <tt>g16run</tt>). | |||
# Gaussian 16 has improved parallel performance on larger numbers of processors. If you want to increase the number of processes for a calculation you should also increase memory by the same factor (if possible). | |||
# Gaussian 16 allows you to pin processes to compute cores which should also increase parallel performance (see [http://gaussian.com/relnotes/?tabid=3 Parallel Performance] for details). If you encounter problems when use pinning you should try to reserve all cores of a complete node. | |||
# Parallelization across nodes with Linda is not possible with Gaussian 16 (no license for Linda). In the partition <tt>mpcp.p</tt> you can use up to 40 compute cores. | |||
# Gaussian 16 supports the use of GPUs for acceleration, however only for K40 and K80 cards. The P100-GPUs available in the cluster are not (yet) supported. | |||
== Documentation == | == Documentation == | ||
For further informations, visit the official homepage of [http://gaussian.com/ Gaussian]. The documentation for the latest version of Gaussian is available from [http://gaussian.com/man/]. | |||
Latest revision as of 15:29, 4 February 2020
Introduction
Gaussian is a computer program for computational chemistry initially released in 1970 by John Pople and his research group at Carnegie Mellon University as Gaussian 70. It has been continuously updated since then. The name originates from Pople's use of Gaussian orbitals to speed up calculations compared to those using Slater-type orbitals, a choice made to improve performance on the limited computing capacities of then-current computer hardware for Hartree–Fock calculations.
Installed version
These currently installed versions are available...
... on envirnoment hpc-uniol-env:
gaussian/g09.b01 gaussian/g09.d01 gaussian/g16.a03 gaussian/g16.C01
... on environment hpc-env/6.4:
Gaussian/g16.a03-AVX2 gaussian/g16.a03 gaussian/g16.c01
We recommend using the environment hpc-env/6.4 as in general more recent software versions are available. If you also want to use GaussView you may have to as the required graphics libraries are not available otherwise.
Available abilities
According to the official homepage, Gaussian 09 Rev. D01 has the following abilities:
- Molecular mechanics
- AMBER
- Universal force field (UFF)
- Dreiding force field
- Semi-empirical quantum chemistry method calculations
- Austin Model 1 (AM1), PM3, CNDO, INDO, MINDO/3, MNDO
- Self-consistent field (SCF methods)
- Hartree–Fock method: restricted, unrestricted, and restricted open-shell.
- Møller–Plesset perturbation theory (MP2, MP3, MP4, MP5).
- Built-in density functional theory (DFT) methods
- B3LYP and other hybrid functionals
- Exchange functionals: PBE, MPW, PW91, Slater, X-alpha, Gill96, TPSS.
- Correlation functionals: PBE, TPSS, VWN, PW91, LYP, PL, P86, B95
- ONIOM (QM/MM method) up to three layers
- Complete active space (CAS) and multi-configurational self-consistent field calculations
- Coupled cluster calculations
- Quadratic configuration interaction (QCI) methods
- Quantum chemistry composite methods (CBS-QB3, CBS-4, CBS-Q, CBS-Q/APNO, G1, G2, G3, W1 high-accuracy methods)
Note: Rev. D.01 has a bug that can cause Gaussian jobs to fail during geometry optimization. See below (or click here) for details and possible work-arounds.
Using Gaussian on the HPC cluster
If you want to find out more about Gaussian on the HPC cluster, you can use the command
module spider gaussian
which will give you an output looking like this:
----- /cm/shared/uniol/modulefiles/chem ----- ... gaussian/g09.b01 ...
To load a specific version of Gaussian use the full name of the module, e.g. to load Gaussian 09 Rev. D.01 (this is also the default):
[abcd1234@hpcl001 ~]$ module load gaussian/g09.d01 [abcd1234@hpcl001 ~]$ module list Currently loaded modules: ... gaussian/g09.d01 ...
Single-node (multi-threaded) jobs
Example: The following job script run_gaussian.job defines a single-node job named g09test on the partition carl.p, using 8 cpus (cores), a total of 6 GB of RAM and 100G of local storage ($TMPDIR) for a maximum run time of 2 hours:
#!/bin/bash #SBATCH --job-name=g09test # job name #SBATCH --partition=carl.p # partition #SBATCH --time=0-2:00:00 # wallclock time d-hh:mm:ss #SBATCH --ntasks=1 # only one task for G09 #SBATCH --cpus-per-task=8 # multiple cpus per task #SBATCH --mem=6gb # total memory per node #SBATCH --gres=tmpdir:100G # reserve 100G on local /scratch INPUTFILE=dimer # load module for Gaussian version to be used (here g09.d01) module load gaussian/g09.d01 # call g09run which now takes up to two arguments # <inputfile>: name of the inputfile (required) # [setnproc]: control whether %NProcShared= is set to match # the requested number of CPUs above (optional) # 0 - do not change the input file (default) # 1 - modify %NProcShared=N if N is larger than # number of CPUs per task requested # 2 - modify %NProcShared=N if N is not equal # to the number of CPUs per task requested g09run $INPUTFILE 2
The example job script and the used input file can be downloaded here (.zip-file).
To submit this job, you have to use the following command:
[abcd1234@hpcl001 ~]$ sbatch run_gaussian.job Submitted batch job 54321
The job will start running as soon as the requested ressources can be allocated. The output of the job (including a checkpoint file in $HOME/g09/CHK) will be produced in the normal directories.
Note: The above procedure and the script are the same for Gaussian 16, except that g09run has to be replaced by g16run. Please read the release notes to find out what has changed in Gaussian 16.
Explanations for the job script:
The expected run time of a job is requested with the --time option e.g. in the format d-hh:mm:ss (you can also omit the :ss part). The default is 2 hours if nothing else is specified in the job script or at job submission.
On the old cluster, you had to request a parallel environment by adding a line ' #$ -pe smp 12' to your job script for a single node job. SLURM does not have parallel environments but you can request CPUs (cores) per task to the same effect. The following settings in the above job script serve as an example:
#SBATCH --partition=carl.p #SBATCH --ntasks=1 #SBATCH --cpus-per-task=8
The maximum amount of "cpus per task" varies for the different partitions (node types) are:
- carl.p = max. 24 cpus per task
- mpcl.p = max. 24 cpus per task
- mpcs.p = max. 24 cpus per task
- mpcb.p = max. 16 cpus per task
- mpcp.p = max. 40 cpus per task
So, if you want to run a single node job on a MPC-BIG node using all available cores, the lines above should be changed to:
#SBATCH --partition=mpcb.p #SBATCH --ntasks=1 #SBATCH --cpus-per-task=16
Note: The numbers mentioned above are the maximum amount of CPUs for each node. You can use them but its not always profitable to do so. It is always a good idea to make some tests to find the optimal number of cores. An example is shown in this diagramm which shows the real (wall clock) and user (CPU) time as a function of number of CPUs (cores).
In the example, memory is requested per node with the --mem-option. This is more practical for Gaussian jobs than requesting memory per core which is also possible with the --mem-per-cpu-option.
Every node has additional local storage which can be used while your job is running. The following line in the example job script
#SBATCH --gres:tmpdir:100G
requests the use of 100GB of local storage for your job. The path for this space is $TMPDIR as always. Further informations can be found here.
Memory Considerations
With the line
#SBATCH --mem 6Gb
the total amount of memory reserved for the job is 6GB (alternative, use --mem-per-cpu to allocate memory per CPU core requested). In the partition carl.p you can reserve up to 117GB per node. You can find out the limits for other partitions using
scontrol show partition <partname.p>
and look for the parameter MaxMemPerNode (the value is in MB). Note, that nodes can be shared between different jobs and the smaller your memory limit is, so more likely your job can run on shared node. Note: if you are not requesting memory explicitly, your job will receive a default amount per requester CPU core. In many cases this may be sufficient and if you are using all cores on a node you also get all available memory.
The Gaussian input files also contain, e.g., the following line in the link 0 section:
%Mem=21000MB
to indicate to Gaussian the available memory.
Important: Memory management is critical for the performance of Gaussian jobs. Which parameter values are optimal is highly dependent on the type of the calculation, the system size, and other factors. Therefore, optimizing your Gaussian job with respect to memory allocation almost always requires (besides experience) some trial and error. The following general remarks may be useful:
In the above example, we have told Gaussian to use almost 21GB (divide MB by 1024) for the job, which means we need to request at least that much for job as well. In fact, since the G09 executables are rather large and have to be resident in memory, there should be some margin (at least 1GB, better 10%, therefore for the above link 0 command we would use --mem 23Gb). This is usually a good choice for DFT calculations.
For MP2 calculations, on the other hand, Gaussian requests about twice the amount of memory specified by the Mem=... directive. If it exceeds the memory reserved for the job, the scheduler will terminate the job. Therefore, for a MP2 calculation the link 0 line should read:
%Mem=11000MB
if 23GB have been requested for the job.
Linda Jobs
Gaussian multi-node jobs (Linda-jobs) can be submitted by simply changing the "--ntasks"-value to something > 1, e.g.
#SBATCH --ntasks=2
Everything else will be handled by the g09run-script, you must no longer add "%LindaWorkers=" to your inputfile!
Note: Linda is only available for Gaussian 09. Gaussian 16 is limited to single node jobs (up to 40 cores in the partition mpcp.p).
Letting g09run set the number of processes
If you have used Gaussian on the old cluster, you will already know what the command "g09run" in your jobscript does: its runs Gaussian with a given input file. This remains unchanged on the new cluster. On the new cluster CARL a feature has been added to g09run. The new feature is simple, but very effective for saving resources that would otherwise be "wasted". We have implemented the possibility to add another option beside naming the input file to "g09run"
setnproc has three different options:
- 0: do not change the input file (default)
- 1: modify %NProcShared=N if N is larger than number of CPUs per task requested
- 2: modify %NProcShared=N if N is not equal to the number of CPUs per task requested
The full syntax of the "g09run"-command looks like this:
g09run <INPUTFILE> [SETNPROC]
where the first argument is mandatory and second optional (i.e. if you do not want to use the new feature you can ignore it).
Example: Using
g09run my_input_file 1
in your job script will cause g09run to check whether %NProcShared is too large for the requested number of CPUs and modify it if needed.
Optimizing the runtime of your jobs
Optimizing the runtime of your jobs will not only safe your time, it will also safe resources (cores, memory, time etc.) for everyone else using the cluster. Therefore you should determine the best amount of cores for your particular job. The following diagram will show the time difference (splitted into real (wall clock) time and user (CPU) time) in terms of the amount of used cores/CPUs. The job we used to gather these times is the example job mentioned above.
(Every test job was done with 6 GB of RAM, except the one with 48 cores which was done with 12 GB of RAM)
As you see in the diagram, increasing the amount of cores will reduce the time your job needs to finish (= real time). However, if you reach a certain amount of cores (16 cores in this example), adding cores will not significantly decrease the real time, it will just increase the time your job is processed by the cpus (= user time). The user time peaks at about 16 cores too, so adding even more cores is not beneficial in any ways.
With Gaussian 16 a Link 0-option has been added that allows you to bind the processes to individual CPU-cores with the aim to improve performance. Binding processes to a core has the advantage that memory access from that process may be more efficient. This can be achieved by replacing the Link 0-line
%nprocshared=24
with the new option
%CPU=0-23
Now Gaussian will at runtime bind each parallel process to a single core rather than letting the OS decide which process to run on which core. In this example, a complete node with 24 cores must be requested.
If in the job script you are requesting less cores than available per node, e.g.
#SBATCH --ntasks=1 # only one task for G16 #SBATCH --cpus-per-task=8 # multiple cpus per task
only 8 cores in this example, than the problem is is that you cannot know in advance which cores will be assigned to you. However, writing
%CPU=0-7
will tell Gaussian to use only the cores with the core IDs 0 to 7. If you get assigned cores with IDs 2,4,5-8,10-11 the above Link 0-option will cause the following error
Error: MP_BLIST has an invalid value
indicating that Gaussian tried to bind a process to a core without having the permission to do so. You Link 0-option needs to be
%CPU=2,4,5-8,10-11
which obviously requires you to modify your input file at runtime. Here is an example how to:
INPUTFILE=dimer
# update inputfile for CPUs
CPUS=$(taskset -cp $$ | awk -F':' '{print $2}')
sed -i -e "/^.*%cpu/I s/=.*$/=$CPUS/" $INPUTFILE
This assumes your inputfile already has a line starting with %CPU= and uses the taskset command to find out, which cores are available for your job.
Current issues with Gaussian 09 Rev. D.01
Currently Rev. D.01 has a bug which can cause geometry optimizations to fail. If this happens, the following error message will appear in your log- or out-file:
Operation on file out of range. FileIO: IOper= 1 IFilNo(1)= -526 Len= 784996 IPos= 0 Q= 46912509105480 .... Error termination in NtrErr: NtrErr Called from FileIO.
The explanation from Gaussian's technical support:
This problem appears in cases where one ends up with different orthonormal subsets of basis functions at different geometries. The "Operation on file out of range" message appears right after the program tries to do an interpolation of two SCF solutions when generating the initial orbital guess for the current geometry optimization point. The goal here is to generate an improved guess for the current point but it failed. The interpolation of the previous two SCF solutions to generate the new initial guess was a new feature introduced in G09 rev. D.01. The reason why this failed in this particular case is because the total number of independent basis functions is different between the two sets of orbitals. We will have this bug fixed in a future Gaussian release, so the guess interpolation works even if the number of independent basis functions is different.
There a number of suggestions from the technical support on how to work around this problem:
A) Use “Guess=Always” to turn off this guess interpolation feature. Option "A" would work in many cases, although it may not be a viable alternative in cases where the desired SCF solution is difficult to get from the default guess and one has to prepare a special initial guess. You may try this for your case.
B) Just start a new geometry optimization from that final point reading the geometry from the checkpoint file. Option "B" should work just fine although you may run into the same issue again if, after a few geometry optimization steps, one ends up again in the scenario of having two geometries with two different numbers of basis functions.
C) Lower the threshold for selecting the linearly independent set by one order of magnitude, which may result in keeping all functions. The aforementioned threshold is controlled by "IOp(3/59=N)" which sets the threshold to 1.0D-N (N=6 is the default). Note that because an IOp is being used, one would need to run the optimization and frequency calculations separately and not as a compound job ("Opt Freq"), because IOps are not passed down to compound jobs. You may also want to use “Integral=(Acc2E=11)” or “Integral=(Acc2E=12)” if you lower this threshold as the calculations may not be as numerically robust as with the default thresholds. Option "C" may work well in many cases where there is only one (or very few) eigenvalue of the overlap matrix that is near the threshold for linear dependencies, so it may just work fine to use "IOp(3/59=7)", which will be keeping all the functions. Because of this situation, and because of potential convergence issues derived from including functions that are nearly linearly dependent, I strongly recommend using a better integral accuracy than the default, for example "Integral=(Acc2E=12)", which is two orders of magnitude better than default.
D) Use fewer diffuse functions or a better balanced basis set, so there aren’t linear dependencies with the default threshold and thus no functions are dropped. Option "D" is good since it would avoid issues with linear dependencies altogether, although it has the disadvantage that you would not be able to reproduce other results with the basis set that you are using.
Changes from G09 to G16
Here is an overview of changes between G09 and G16 in general and regarding the use of the cluster:
- Changes of the latest version of Gaussian can always be found in the release notes.
- Gaussian 16 can be used on the cluster by loading the corresponding module and using g16run within the job script.
- Link 0 (%-lines) inputs can now be specified in different ways (see Link 0 Equivs.), which in principle should work on the cluster as well (with the exception of command-line arguments if you use g16run).
- Gaussian 16 has improved parallel performance on larger numbers of processors. If you want to increase the number of processes for a calculation you should also increase memory by the same factor (if possible).
- Gaussian 16 allows you to pin processes to compute cores which should also increase parallel performance (see Parallel Performance for details). If you encounter problems when use pinning you should try to reserve all cores of a complete node.
- Parallelization across nodes with Linda is not possible with Gaussian 16 (no license for Linda). In the partition mpcp.p you can use up to 40 compute cores.
- Gaussian 16 supports the use of GPUs for acceleration, however only for K40 and K80 cards. The P100-GPUs available in the cluster are not (yet) supported.
Documentation
For further informations, visit the official homepage of Gaussian. The documentation for the latest version of Gaussian is available from [1].