Difference between revisions of "Matlab Examples using MDCS"
| (10 intermediate revisions by one other user not shown) | |||
| Line 763: | Line 763: | ||
First, consider the small example code | First, consider the small example code | ||
for n=1:4 | |||
for n=1:4 | n | ||
end | |||
end | |||
contained in the .m-file <tt>test_output.m</tt> and sent to the scheduler via | contained in the .m-file <tt>test_output.m</tt> and sent to the scheduler via | ||
sched = findResource('scheduler','Configuration','HERO'); | |||
sched = findResource('scheduler','Configuration','HERO'); | job = batch(sched,'test_output'); | ||
job = batch(sched,'test_output'); | After termination the job will have written the numbers 1 through 4 to the | ||
After termination the job will have written the | |||
standard output. | standard output. | ||
| Line 780: | Line 776: | ||
#1 Use the <tt>diary</tt> as you would using your local MATLAB installation | #1 Use the <tt>diary</tt> as you would using your local MATLAB installation | ||
#2 Look up the job related auxiliary files in the <tt>jobData</tt> folder on your local desktop computer. | #2 Look up the job related auxiliary files in the <tt>jobData</tt> folder on your local desktop computer. | ||
Both options will be illustrated below. | Both options will be illustrated below (NOTE: this section was motivated by the user <tt>gepp0026</tt>). | ||
=== Using the <tt>diary</tt> option === | === Using the <tt>diary</tt> option === | ||
| Line 829: | Line 825: | ||
</nowiki> | </nowiki> | ||
The first part is actually a warning message that stems from the client and host being physically separated (i.e. not on the same file system) and not harmful. You can ignore | The first part is actually a warning message that stems from the client and host being physically separated (i.e. not on the same file system) and not harmful. You can ignore it without worries. The second part lists the command window output of the small application, i.e. the state of the variable <tt>n</tt> throughout the different iterations. | ||
=== Looking up the auxiliary job information files === | === Looking up the auxiliary job information files === | ||
| Line 840: | Line 836: | ||
<tt>jobData/Job136</tt> you can find a wealth of job related auxiliary files. | <tt>jobData/Job136</tt> you can find a wealth of job related auxiliary files. | ||
I don't want to go through them in detail, but here is a list to give you an | I don't want to go through them in detail, but here is a list to give you an | ||
idea what you should expect to find for a job that employs | idea what you should expect to find for a job that employs a single worker: | ||
<nowiki> | <nowiki> | ||
distributedJobWrapper.sh | distributedJobWrapper.sh | ||
| Line 862: | Line 858: | ||
workspace (with there respective size and data types) via the command | workspace (with there respective size and data types) via the command | ||
whos | whos | ||
which, for the above application, yields (if you open up a new MATLAB session) | which, for the above application, yields (if you open up a new MATLAB session for this puropose) | ||
<nowiki> | <nowiki> | ||
Name Size Bytes Class Attributes | Name Size Bytes Class Attributes | ||
| Line 913: | Line 909: | ||
4 | 4 | ||
</ | </nowiki> | ||
Again, the first part is a warning message that stems from the client and host being physically separated (i.e. not on the same file system) and not harmful. You can ignore | Again, the first part is a warning message that stems from the client and host being physically separated (i.e. not on the same file system) and not harmful. You can ignore it without worries. The second part lists the command window output of the small application, i.e. the state of the variable <tt>n</tt> throughout the different iterations. | ||
== Parallelization using parfor loops == | |||
Probably the most simple way to parallelize already written code is by using | |||
<tt>parfor</tt> loops. As its name suggests, | |||
the command <tt>parfor</tt> indicates a <tt>for</tt> loop which is executed in parallel. | |||
As soon as your MATLAB session encounters a <tt>parfor</tt> loop, | |||
the range of the iteration variable is subdivided into a specified number of blocks which | |||
are in turn processed by different workers. | |||
Note that <tt>parfor</tt> loops require the iterations to be '''independent''' of each other | |||
since it is in general not possible to ensure that the iterations finish in a particular order. | |||
=== Basic example: Vector-Matrix multiplication === | |||
To facilitate intuition, consider a vector-matrix multiplication, for given <math>N\times 1</math>-vector <math>b</math> and <math>N\times N</math>-matrix <math>A</math>, as one would do it by hand: | |||
<math>c_i=\sum_j A_{ij}b_j</math>. Or, in terms of a (arguably non-optimal) sequential MATLAB snippet: | |||
<nowiki> | |||
for i=1:N | |||
tmp=0.; | |||
for j=1:N | |||
tmp=tmp+A(i,j)*b(j); | |||
end | |||
c(i)=tmp; | |||
end | |||
</nowiki> | |||
As evident from that code snipped, the different entries in the solution vector | |||
<math>c</math> do not depend on each other. In principle, each entry of the solution | |||
vector might be computed on a different worker, even at the same time, without affecting the | |||
results. | |||
Hence, this piece of code might be amended by using a <tt>parfor</tt> loops instead. A complete example | |||
using <tt>parfor</tt> reads: | |||
<nowiki> | |||
N=2048; | |||
b=rand(N,1); | |||
A=rand(N,N); | |||
parfor i=1:N | |||
tmp=0.; | |||
for j=1:N | |||
tmp=tmp+A(i,j)*b(j); | |||
end | |||
c(i)=tmp; | |||
end | |||
</nowiki> | |||
In order to submit this example to the HPC system, you need to first save the | |||
example as .m-file, e.g. <tt>myExample_parfor.m</tt>, and then submit it via | |||
<nowiki> | |||
sched=findResources('scheduler','Configuration','HERO'); | |||
job=batch(sched,'myExample_parfor','matlabpool',8); | |||
</nowiki> | |||
=== Variable slicing === | |||
MATLAB has a particular classification scheme for variables within <tt>parfor</tt> loops (see [http://www.mathworks.de/de/help/distcomp/advanced-topics.html|here]). | |||
A variable might, e.g. | |||
be a ''loop variable'', as the variable <tt>i</tt> in the above example, a ''temporary'' | |||
variable, as the variable <tt>tmp</tt> above, or a ''sliced'' variable (note that there | |||
are more possible classifications). | |||
A sliced variable refers to an array whose segments are operated on by | |||
different iterations of the loop. Just like for the entries of the output array | |||
<tt>c</tt>. Note that the same holds also for the rows of the input Matrix <tt>A</tt>: | |||
to compute the entry <tt>c(i)</tt> only the row <tt>A(i,:)</tt> is needed. In this sense, <tt>c(i)</tt> is a sliced ''output'' variable, while <tt>A(i,:)</tt> might be used as a sliced ''input'' variable. | |||
A sliced version of the above example reads: | |||
<nowiki> | |||
N=2048; | |||
b=rand(N,1); | |||
A=rand(N,N); | |||
parfor i=1:N | |||
c(i)=A(i,:)*b(:); | |||
end | |||
</nowiki> | |||
Note that <tt>parfor</tt> | |||
loops start by first distributing data to the allocate workers. By using sliced | |||
variables only the data which is really needed by a particular worker is sent to it. | |||
Hence, the use of sliced variables might result in less overall memory and/or time consumption. | |||
=== Examples where parfor loops do not apply === | |||
[http://en.wikipedia.org/wiki/Recurrence_relation| Recurrence relations], i.e. equations that define a sequence recursively are, by their very nature, not parallelizable. | |||
=== Further considerations === | |||
In principle, by encountering a <tt>parlor</tt> loop there is some overhead | |||
associated to the startup of parallel processes and to the distribution of data | |||
to the workers. Hence, it might not be a good advice to simply replace each | |||
loop by a parfor loop. E.g. for the intuitive, yet trivial vector-matrix | |||
multiplication example the use of a <tt>parlor</tt> loop might not pay off, | |||
since MATLAB offers efficient implementations for that purpose. Often it pays | |||
off to first search for the function that is responsible for the majority of | |||
execution time and to parallelize that. | |||
== Other Topics == | == Other Topics == | ||
=== MDCS working mode === | |||
On the local HPC facilities we use MATLAB for a non-interactive processing of jobs, i.e. in batch mode, only. | |||
The scheduler (we use SGE) offers the dedicated <tt>mdcs</tt> parallel environment which is used to run a specified | |||
number of workers as applications. These applications are started to process user-supplied tasks, and they | |||
stop as soon as the tasks are completed. More information on this working mode might be found | |||
[http://www.mathworks.de/products/parallel-computing/description8.html here]. | |||
=== The parallel environment memory issue === | === The parallel environment memory issue === | ||
Latest revision as of 10:44, 6 July 2016
A few examples for Matlab applications using MDCS (prepared using Matlab version R2011b) are illustrated below.
Example application: 2D random walk
Consider the Matlab .m-file myExample_2DRandWalk.m (listed below), which among other things illustrates the use of sliced variables and independent stremas of random numbers for use with parfor-loops.
This example program generates a number of N independent
2D random walks (a single step has steplength 1 and a
random direction). Each random walk performs tMax steps.
At each step t, the radius of gyration (Rgyr) of walk i
is stored in the array Rgyr_t in the entry Rgyr_t(i,t).
While the whole data is availabe for further postprocessing,
only the average radius of gyration Rgyr_av and the respective
standard error Rgyr_sErr for the time steps 1...tMax are
computed immediately (below it will also be shown how to store the data in an output file on HERO
for further postprocessing).
%% FILE: myExample_2DRandWalk.m
% BRIEF: illustrate sliced variables and independent streams
% of random numbers for use with parfor-loops
%
% DEPENDENCIES:
% singleRandWalk.m - implements single random walk
% averageRgyr.m - computes average radius of gyration
% for time steps 1...tMax
%
% AUTHOR: Oliver Melchert
% DATE: 2013-06-05
%
N = 10000; % number of independent walks
tMax = 100; % number of steps in individual walk
Rgyr_t = zeros(N,tMax); % matrix to hold results: row=radius
% of gyration as fct of time;
% col=independent random walk instances
parfor n=1:N
% create random number stream seeded by the
% current value of n; you can obtain a list
% of all possible random number streams by
% typing RandStream.list in the command window
myStream = RandStream('mt19937ar','Seed',n);
% obtain radius of gyration as fct of time for
% different independent random walks (indepence
% of RWs is ensured by connsidering different
% random number streams for each RW instance)
Rgyr_t(n,:) = singleRandWalk(myStream,tMax);
end
% compute average Rgyr and its standard error for all steps
[Rgyr_av,Rgyr_sErr] = averageRgyr(Rgyr_t);
As liste above, the .m-file depends on the following files:
- singleRandWalk.m, implementing a single random walk, reading:
function [Rgyr_t]=singleRandWalk(randStream,tMax)
% Usage: [Rgyr_t]=singleRandWalk(randStream,tMax)
% Input:
% randStream - random number stream
% tMax - number of steps in random walk
% Output:
% Rgyr_r - array holding the radius of gyration
% for all considered time steps
x=0.;y=0.; % initial walker position
Rgyr_t = zeros(tMax,1);
for t = 1:tMax
% implement random step
phi=2.*pi*rand(randStream);
x = x+cos(phi);
y = y+sin(phi);
% record radius of gyration for current time
Rgyr_t(t)=sqrt(x*x+y*y);
end
end
- averageRgyr.m, which computes the average radius of gyration of the random walks for time steps
1...tMax, reading:
function [avList,stdErrList]=averageRgyr(rawDat)
% Usage: [av]=averageRgyr(rawDat)
% Input:
% rawData - array of size [N,tMax] where N is the
% number of independent random walks and
% tMax is the number of steps taken by an
% individual walk
% Returns:
% av - aveage radius of gyration for the steps
[Lx,Ly]=size(rawDat);
avList = zeros(Ly,1);
stdErrList = zeros(Ly,1);
for i = 1:Ly
[av,var,stdErr] = basicStats(rawDat(:,i));
avList(i) = av;
stdErrList(i) = stdErr;
end
end
function [av,var,stdErr]=basicStats(x)
% usage: [av,var,stdErr]=basicStats(x)
% Input:
% x - list of numbers
% Returns:
% av - average
% var - variance
% stdErr - standard error
av=sum(x)/length(x);
var=sum((x-av).^2)/(length(x)-1);
stdErr=sqrt(var/length(x));
end
For test purposes one might execute the myExample_2DRandWalk.m directly from within a Matlab session on a local Desktop PC.
Specifying file dependencies
So as to sumbit the respective job to the local HPC system one might assemble the following job submission script, called mySubmitScript_v1.m:
sched = findResource('scheduler','Configuration','HERO');
jobRW =...
batch(...
sched,...
'myExample_2DRandWalk',...
'matlabpool',2,...
'FileDependencies',{...
'singleRandWalk.m',...
'averageRgyr.m'...
}...
);
In the above job submission script, all dependent files are listed as FileDependencies. I.e., the .m-files specified therein are copied from your local desktop PC to the HPC system at run time.
Now, from within a Matlab session I navigate to the Folder where the above .m-files are located in and call the job submission script, i.e.:
>> cd MATLAB/R2011b/example/myExamples_matlab/RandWalk/ >> mySubmitScript_v1 runtime = 24:0:0 (default) memory = 1500M (default) diskspace = 50G (default)
before the job is actually submitted, I need to specify my user ID and password, of course. Once the job is successfully submitted, I can check the state of the job via typing jobRW.state. However, if you want to get some more information on the status of your job, you might want to try to log-in on the HPC system and to simply type the command qstat on the commandline. This will yield several details related to your job which you might process further to see on which execution nodes your job runs, why it won't start directly etc. Note that MATLAB provides only a wrapper for the qstat command which in some cases result in a misleading output. E.g., if, for some reason, your job changes to the error-state it might be that MATLAB erroneously reports it to be in the finished-state.
Once the job (really) has finished, i.e.,
>> jobRW.state ans = finished
I might go on and load the results to my desktop computer, giving
>> res=load(jobRW);
>> res
res =
N: 10000
Rgyr_av: [100x1 double]
Rgyr_sErr: [100x1 double]
Rgyr_t: [10000x100 double]
ans: 'finished'
res: [1x1 struct]
tMax: 100
However, note that there are several drawbacks related to the usage of FileDependencies: E.g.,
- each worker gets an own copy of the respective .m-files when the job starts (in particular, workers that participate in the computing process do not share a set of .m-files in a common location),
- the respective .m-files are not available on the HPC system once the job has finished,
- comparatively large input files need to be copied to the HPC system over and over again, if several computations on the same set of input data are performed.
In many cases a different procedure, based on specifying PathDependencies, outlined below in detail, might be recommendend.
Specifying path dependencies
Basically there are two ways to specify path dependencies. You might either specify them in your job submission script or directly in your main MATLAB .m-file. Below, both approaches are illustrated.
Modifying the job submission script
The idea underlying the specification of path dependencies is that there might be MATLAB modules (or sets of data) you want to routinely use over and over again. Then, having these modules available on your local desktop computer and using FileDependencies to copy them to the HPC system at run time results in time and memory consuming, unnecessary operations.
As a remedy you might adopt the following two-step procedure:
- copy the respective modules to the HPC system
- upon submitting the job from your local desktop pc, indicate by means of the key word PathDependencies where (on HERO) the respective data files can be found.
This eliminates the need to copy the respective files using the FileDependencies statement. An example of how to accomplish this for the random walk example above is given below.
Just for arguments, say, the content of the two module files singleRandWalk.m and averageRgyr.m will not be changed in the near future and you want to use both files on a regular basis when you submit jobs to the HPC cluster. Hence, it would make sense to copy them to the HPC system and to specify within your job submission script where they can be found for use by any execution host. Following the above two steps you might proceed as follows:
1. create a folder where you will copy both files to. To facilitate intuition and to make this as explicit as possible, I created all folders along the path
/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/random_walk
and copied both files there, see
alxo9476@hero01:~/SIM/MATLAB/R2011b/my_modules/random_walk$ ls averageRgyr.m singleRandWalk.m
2. Now, it is no more necessary to specify both files as file dependencies as in the example above. Instead, in your job submission file (here called mySumbitScript_v2.m) you might now specify a path depenency (which relates to your filesystem on the HPC system) as follows:
sched = findResource('scheduler','Configuration','HERO');
jobRW =...
batch(...
sched,...
'myExample_2DRandWalk',...
'matlabpool',2,...
'PathDependencies',{'/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/random_walk'}...
);
This has the benefit that the files will not be copied to the HPC system at run time and that there is only a single copy of those files on the HPC system, which can be used by all execution hosts (so no multiple copies of the same files necessarry as with the use of file dependencies).
Again, from within a Matlab session navigate to the folder where the job submission file and the main file myExample_2DRandWalk.m are located in and call the job submission script. For me, this reads:
>> cd MATLAB/R2011b/example/myExamples_matlab/RandWalk/ >> mySubmitScript_v2 runtime = 24:0:0 (default) memory = 1500M (default) diskspace = 50G (default)
again, before the job is actually submitted, I need to specify my user ID and password (since I started a new MATLAB session in between). Once the job has finished I might go on and load the results to my desktop computer, giving
>> res=load(jobRW);
>> res
res =
N: 10000
Rgyr_av: [100x1 double]
Rgyr_sErr: [100x1 double]
Rgyr_t: [10000x100 double]
ans: 'finished'
res: [1x1 struct]
tMax: 100
Modifying the main m-file
As an alternative to the above procedure, you might add the folder
/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/
to your local MATLAB path by adding the single line
addpath(genpath('/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/'));
to the very beginning of the file myExample_2DRandWalk.m. For completeness, note that this adds the above folder my_modules and all its subfolders to your MATLAB path. Consequently, all the m-files contained therein will be available to the execution nodes which contribute to a MATLAB session. Further note that it specifies an absolute path, referring to a location within you filesystem on the HPC system.
In this case, a proper job submission script (mySubmitScript_v3.m) reads simply:
sched = findResource('scheduler','Configuration','HERO');
jobRW =...
batch(...
sched,...
'myExample_2DRandWalk',...
'matlabpool',2...
);
As a personal comment, note that, from a point of view of submitting a job to the HPC System, I would always prefer the explicit way of stating path dependencies in the job submission file over the implicit way of indirectly implying them using a modification of the main m-file. The latter choice seems much more vulnerable to later changes!
Storing data on HERO
Consider a situation where your application produces lots of data you want to store for further postprocessing. Often, in particular when you produce lots of data, you also don't want the data to be copied back to your desktop computer, immediately. Instead you might want to store the data on HERO and perhaps mount your HPC-homedirectory later to relocate the data (or the like). Note that, as a default HERO user, you have 110GB of disk space available under your home directory. Below it is illustrated, by means of the random walk example introduced earlier, how to store output data on the HPC system.
The only thing you need to do is to specify a path within your main m-file under which the data should be stored. Therefore you first have to create the corresponding sequence of folders if they do not exist already. To facilitate intuition: in my case I decided to store the data under the path
/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_data/random_walk/
I created the folder my_data/rand_walk/ for that purpose. Right now, the folder is empty.
In the main file (here called myExample_2DRandWalk_saveData.m) I implemented the following changes:
N = 10000; % number of independent walks
tMax = 100; % number of steps in individual walk
Rgyr_t = zeros(N,tMax); % matrix to hold results: row=radius
% of gyration as fct of time;
% col=independent random walk instances
% absolute path to file on HERO where data will be saved
outFileName=sprintf('/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_data/random_walk/rw2d_N%d_t%d.dat',N,tMax);
parfor n=1:N
% create random number stream seeded by the
% current value of n; you can obtain a list
% of all possible random number streams by
% typing RandStream.list in the command window
myStream = RandStream('mt19937ar','Seed',n);
% obtain radius of gyration as fct of time for
% different independent random walks (indepence
% of RWs is ensured by connsidering different
% random number streams for each RW instance)
Rgyr_t(n,:) = singleRandWalk(myStream,tMax);
end
% compute average Rgyr and its standard error for all steps
[Rgyr_av,Rgyr_sErr] = averageRgyr(Rgyr_t);
% write data to output file on HERO
saveData_Rgyr(outFileName,Rgyr_av, Rgyr_sErr);
Note that the outFileName is specified directly in the main m-file (there are also more elegant ways to accomplish this, however, for the moment this will do!) and the new function, termed saveData_Rgyr is called. The latter function will just write out some statistical summary measures related to the gyration radii of the 2D random walks. For completeness, it reads:
function saveData(fileName,Rgyr_av,Rgyr_sErr)
% Usage: saveData(fileName,myData)
% Input:
% fileName - name of output file
% myData - data to be saved in file
% Returns: nothing
outFile = fopen(fileName,'w');
fprintf(outFile,'# t Rgyr_av Rgyr_sErr \n');
for i = 1:length(Rgyr_av)
fprintf(outFile,'%d %f %f\n',i,Rgyr_av(i),Rgyr_sErr(i));
end
fclose(outFile);
end
Now, say, my opinion on the proper output format is not settled yet and I consider to experiment with different kinds of output formatting styles, for that matter. Then it is completely fine to specify some of the dependent files as path dependencies (namely those that are unlikely to change soon) and others as file dependencies (namely those which are under development). Here, joining in the specification of path dependencies within a job submission script, a proper submission script (here called mySubmitScript_v4.m) might read:
sched = findResource('scheduler','Configuration','HERO');
jobRW =...
batch(...
sched,...
'myExample_2DRandWalk_saveData',...
'matlabpool',2,...
'FileDependencies',{'saveData_Rgyr.m'},...
'PathDependencies',{'/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_modules/random_walk'}...
);
Again, starting a MATLAB session, changing to the directory where the myExample_2DRandWalk_saveData.m and saveData_Rgyr.m files are located in, launching the submission script mySubmitScript_v5.m and waiting for the job to finish, i.e.
>> cd MATLAB/R2011b/example/myExamples_matlab/RandWalk/ >> mySubmitScript_v4 runtime = 24:0:0 (default) memory = 1500M (default) diskspace = 50G (default) >> jobRW.state ans = finished
Once the job is done, the output file rw2d_N10000_t100.dat (as specified in the main script myExample_2DRandWalk_saveData.m) is created in the folder under the path
/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/my_data/random_walk/
which contains the output data in the format as implemented by the function saveData_Rgyr.m, i.e.
alxo9476@hero01:~/SIM/MATLAB/R2011b/my_data/random_walk$ head rw2d_N10000_t100.dat # t Rgyr_av Rgyr_sErr 1 1.000000 0.000000 2 1.273834 0.006120 3 1.572438 0.007235 4 1.795441 0.008645 5 1.998900 0.009714 6 2.168326 0.010867 7 2.355482 0.011766 8 2.519637 0.012721 9 2.683048 0.013456
Again, after the job has finished, the respective file remains on HERO and is available for further postprocessing. It is not automatically copied to your local desktop computer.
Postprocessing of the simulated data
Just for completeness a brief analysis and display of the 2D random walk data is presenter here.
Once the job has finished and the resulting data is loaded to the desktop
computer via res=load(jobRW); its possible to postprocess and display
some of the results via the m-file finalPlot.m which reads:
function finalPlot(res)
% Usage: finalPlot(res)
% Input:
% res - simulation results for 2D rand walks
% Returns: (none)
%
subplot(1,2,1), plotHelper(res);
subplot(1,2,2), histHelper(res);
end
function plotHelper(res)
% Usage: plotHelper(res)
%
% implements plot of the gyration radius as
% function of the walk length
%
% Input:
% res - simulation results for 2D rand walks
% Returns: (none)
%
xRange=1:1:100;
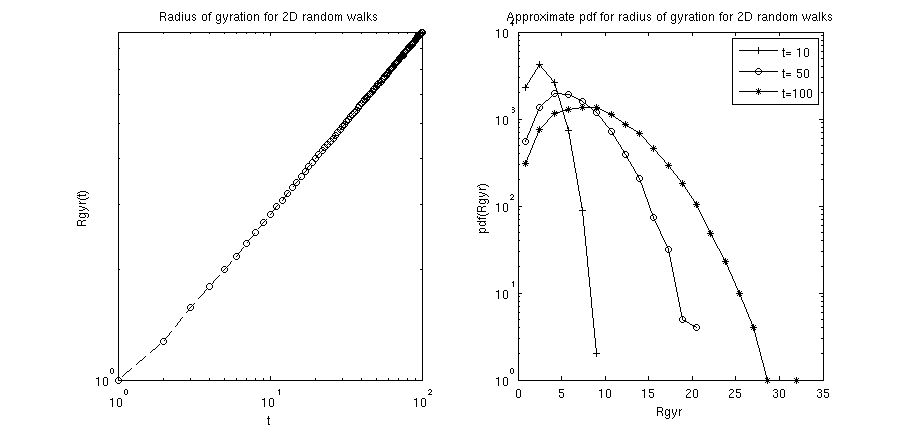
loglog(xRange,res.Rgyr_av,'--ko');
title('Radius of gyration for 2D random walks');
xlabel('t');
ylabel('Rgyr(t)');
end
function histHelper(res)
% Usage: histHelper(res)
%
% implements histogram of the pdf for gyration
% radii at chosen times steps
% of the walk length
%
% Input:
% res - simulation results for 2D rand walks
% Returns: (none)
%
set(0,'DefaultAxesColorOrder',[0 0 0],...
'DefaultAxesLineStyleOrder','-+|-o|-*');
tList=[10,50,100];
[events,key]=hist(res.Rgyr_t(:,tList),20);
semilogy(key,events);
title('Approximate pdf for radius of gyration for 2D random walks');
xlabel('Rgyr');
ylabel('pdf(Rgyr)');
key = legend('t= 10','t= 50','t=100');
end
Therein, the main function calls two subfunctions:
plotHelper(res): implements the gyration radius of the 2D random walks as a function of the number of steps in the walk (i.e. the elapsed time since the walk started). The radius of gyration of such an unhindered 2D random walks generally scales (for not too short times ).histHelper(res): implements histograms of the probability density function (pdf) for the gyration radii at specified time steps. In the above example the time steps and where chosen. For not too short times, one can expect that the pdf is properly characterized by the Rayleigh distribution .
Calling the m-file should yield a figure similar to the following:
Recovering jobs (after closing and restarting MATLAB)
Once you submitted one or several jobs, you might just shut down your local MATLAB session. If you open up a new MATLAB session later on, you can recover the jobs you sent earlier by first getting a connection to the scheduler via
sched = findResource('scheduler','Configuration','HERO');
You can go on and list the jobs in the database using
sched.Jobs
Further, to get a grip on the very first job in the list you might type
myJob_no1 = sched.Jobs(1)
which recovers the respective job and lists some of the job details already.
Monitoring Jobs on the HPC system
Once you submitted your job to the scheduler, in our case the third-party scheduler SGE, you have several options to check the status of your job. Of course, if the job you submitted is called myJob, you could just type
myJob.state
on your MATLAB command window. However, if you want to get some more information on the status of your job, you might want to try to log-in on the HPC system and to simply type the command qstat on the commandline. This will yield several details related to your job which you might process further to see on which execution nodes your job runs, why it won't start directly etc. Note that MATLAB provides only a wrapper for the qstat command which in some cases result in a misleading output. E.g., if, for some reason, your job changes to the error-state it might be that MATLAB erroneously reports it to be in the finished-state. In this regard, loging in on the HPC system might shed some more light on how the state of your job actually is. Consider the following example:
To make this as explicit as possible, I will document here an exemplary job I submitted to the HPC system. In particular, I submitted the above random Walk example using the mySubmitScript_v2.m job-submission script. I submitted the job from my local desktop PC (i.e. the Client), of course, and specified an overall amount of 3 workers to work on the job. As soon as the job is sent to the HPC system, it is handed over to the Scheduler (here, as third-party scheduler we use SGE), which assigns a unique job-Id to the job and places the job in a proper queue. Now, logged in to my HERO account and immediately after having submitted the job, I type
qstat
on the commandline and find
job-ID prior name user state submit/start at queue slots ja-task-ID --------------------------------------------------------------------------------------------------------- 760391 0.00000 Job123 alxo9476 qw 07/08/2013 14:27:26 3
According to this, the job with job-ID 760391 has priority 0.00000, name Job123 and resides in state qw loosely translated to "enqueued and waiting". Also, the above output indicates that the job requires a number of 3 slots, which, in MATLAB terms can be translated to 3 workers. The column for the ja-task-ID, referring to the id of the particular task stemming from the execution of a job array (we don't work through a job array since we submitted a single job), is actually empty and of no importance, here. Soon after, the priority of the job will take a value in between 0.5 and 1.0 (usually only slightly above 0.5), slightly increasing until the job starts. Here, after waiting a few seconds qstat triggers the output
job-ID prior name user state submit/start at queue slots ja-task-ID --------------------------------------------------------------------------------------------------------- 760391 0.50535 Job123 alxo9476 r 07/08/2013 14:28:03 mpc_std_shrt.q@mpcs021 3
indicating that the job is now in state r, i.e. running, and that the so called Master process (which coordinates the 3 workers) is started on the execution host mpcs021. To see where the individual workers are started just type
qstat -g t
to get a more detailed view on the job:
job-ID prior name user state submit/start at queue master ja-task-ID
----------------------------------------------------------------------------------------------------------
760391 0.50535 Job123 alxo9476 r 07/08/2013 14:28:03 mpc_std_shrt.q@mpcs003 SLAVE
mpc_std_shrt.q@mpcs003 SLAVE
760391 0.50535 Job123 alxo9476 r 07/08/2013 14:28:03 mpc_std_shrt.q@mpcs021 MASTER
mpc_std_shrt.q@mpcs021 SLAVE
So, apparently, the job was split across the two execution hosts mpcs021 and mpcs003. Even more information on the job is available if you use qstat with the particular job-Id, i.e. from the query
qstat -j 760391
one obtains a lot of information related to job 760391, here I show just the first few lines related to the job above
============================================================== job_number: 760391 exec_file: job_scripts/760391 submission_time: Mon Jul 8 14:27:26 2013 owner: alxo9476 uid: 59118 group: ifp gid: 12400 sge_o_home: /user/fk5/ifp/agcompphys/alxo9476 sge_o_log_name: alxo9476 sge_o_path: /cm/shared/apps/gcc/4.7.1/bin:/cm/shared/apps/system/bin:/usr/lib64/qt-3.3/bin:/usr/kerberos/bin:/usr/local/bin:/bin:/usr/bin:/sbin:/usr/sbin:/sbin:/usr/sbin:/cm/shared/apps/sge/6.2u5p2/bin/lx26-amd64:/cm/shared/apps/sge/6.2u5p2/local/bin sge_o_shell: /bin/bash sge_o_workdir: /user/fk5/ifp/agcompphys/alxo9476 sge_o_host: hero01 account: sge merge: y hard resource_list: h_rt=86400,h_vmem=1500M,h_fsize=50G mail_list: alxo9476@hero01.cm.cluster notify: FALSE job_name: Job123 stdout_path_list: NONE:NONE:/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/jobData/Job123/Job123.mpiexec.log jobshare: 0 shell_list: NONE:/bin/sh env_list: MDCE_DECODE_FUNCTION=parallel.cluster.generic.parallelDecodeFcn,MDCE_STORAGE_CONSTRUCTOR=makeFileStorageObject,MDCE_JOB_LOCATION=Job123,MDCE_MATLAB_EXE=/cm/shared/apps/matlab/r2011b/bin/worker,MDCE_MATLAB_ARGS=-parallel,MDCE_DEBUG=true,MDCE_STORAGE_LOCATION=/user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/jobData,MDCE_CMR=/cm/shared/apps/matlab/r2011b,MDCE_TOTAL_TASKS=3 script_file: /user/fk5/ifp/agcompphys/alxo9476/SIM/MATLAB/R2011b/jobData/Job123/parallelJobWrapper.sh parallel environment: mdcs range: 3 ...
the rest of the output refers to not so easy to interpret scheduling info that basically summarizes which execution hosts where not considered. From this you can get all kinds of information about the job, e.g. a summary of specified resources (see hard resource_list) and the standard output path to which all output regarding the startup of the workers and the job are send to (see stdout_path_list). As further detail, note that the parallel environment used to process the job is called mdcs, which currently has an overall number of 224 slots (i.e. workers) available.
As soon as the job is finished (either successfully or it failed with some kind of error), it is not possible to retrieve information using qstat. However, for finished jobs you can use the command qacct to obtain some information about the finished jobs. Here, for the above job I type
qacct -j 760391
and obtain
============================================================== qname mpc_std_shrt.q hostname mpcs021.mpinet.cluster group ifp owner alxo9476 project NONE department defaultdepartment jobname Job123 jobnumber 760391 taskid undefined account sge priority 0 qsub_time Mon Jul 8 14:27:26 2013 start_time Mon Jul 8 14:28:05 2013 end_time Mon Jul 8 14:30:12 2013 granted_pe mdcs slots 3 failed 0 exit_status 0 ru_wallclock 127 ru_utime 18.922 ru_stime 3.373 ru_maxrss 384992 ru_ixrss 0 ru_ismrss 0 ru_idrss 0 ru_isrss 0 ru_minflt 232574 ru_majflt 3142 ru_nswap 0 ru_inblock 0 ru_oublock 0 ru_msgsnd 0 ru_msgrcv 0 ru_nsignals 0 ru_nvcsw 53953 ru_nivcsw 9178 cpu 22.296 mem 0.002 io 0.001 iow 0.000 maxvmem 356.406M arid undefined
Among the details listed there might be a few that might help in the process of interpreting what might have happened to your job in case something went wrong. E.g. see the keywords failed and exit_status. Also the maximally consumed memory maxvmem might be of interest in the process of finding errors.
Reading error messages of failed jobs
This evening I submitted a job to the HPC system. During the submission procedure on my local MATLAB session I called it myJob. I submitted it using the standard resource specifications. However, soon after the job started, it attempted to allocate a lot of memory ... more than what was granted by using the standard resources. Needless to say that the job got immediately killed! So, before coming to that conclusion I just knew that the job was finished very early and using the statement
myJob.state
on the MATLAB command window it stated
finished
So, first I was surprised that everything went so fast! However, as discussed also earlier, sometimes the MATLAB output of the myJob.state command can be somewhat misleading and I found the fact that the program finished so fast slightly suspicious. So I proceeded by checking the list of tasks related to the job, i.e. the result of the single master task and the 3 slave tasks that comprise the job. Therefore I wrote the following in the MATLAB command window
>> myJob.Tasks
ans =
Tasks: 4-by-1
=============
# Task ID State FinishTime Function Name Error
-----------------------------------------------------------------------------------------
1 1 finished Jul 08 16:14... @parallel.internal.cluster.executeS... Error
2 2 finished Jul 08 16:14... @distcomp.nop
3 3 finished Jul 08 16:14... @distcomp.nop
4 4 finished Jul 08 16:14... @distcomp.nop
Albeit all 4 tasks where finished, note that task 1 (generally the master task) reports an error which is not specified further. To get some more information on that error, I put that particular task under scrutiny. Therefore I simply specify the particular task I am interested in by typing
>> myJob.Task(1)
ans =
Task ID 1 from Job ID 124 Information
=====================================
State : finished
Function : @parallel.internal.cluster.executeScript
StartTime : Mon Jul 08 16:13:40 CEST 2013
Running Duration : 0 days 0h 0m 39s
- Task Result Properties
ErrorIdentifier : MATLAB:nomem
ErrorMessage :
: Out of memory. Type HELP MEMORY for your options.
:
: Error stack:
: dietz2011interauralfunctions.m at 105
: dietz2011_modaj.m at 395
: feature_extraction.m at 68
: pitch_direction_main.m at 55
Error Stack : parallel_function.m at 598
: pitch_direction_main_run.m at 13
: executeScript.m at 24
From the ErrorMessage it is now evident where the error stems from. The Error Stack even lists a hierarchy of .m files with the precise location the error was encountered at.
Note that if I had tried to load the result of the job in a straight forward manner, I would have received the myJob.Task(1).ErrorMessage as output, see
>> res = load(myJob) Error using distcomp.abstractjob/load (line 64) Error encountered while running the batch job. The error was: Out of memory. Type HELP MEMORY for your options. Error stack: dietz2011interauralfunctions.m at 105 dietz2011_modaj.m at 395 feature_extraction.m at 68 pitch_direction_main.m at 55
As a remedy I restarted the job requesting some more memory by adding the line
set(sched, 'ParallelSubmitFcn',cat(1,sched.ParallelSubmitFcn,'memory','4G'));
to my job submission script.
Note that on HERO one also could have arrived at the conclusion that the job did not finish correctly by simply checking the exit_status of the job, which, in this case, has had the job-Id 760409. Using the qacct tool by means of the command
qacct -j 760409 | grep "exit_status"
it yields
exit_status 123
However, also here, no immediate hint on what caused the error in the first place.
Viewing command window output
Consider the situation where your application writes out several things to the standard output and you want to have a look at the output after the application has terminated.
First, consider the small example code
for n=1:4 n end
contained in the .m-file test_output.m and sent to the scheduler via
sched = findResource('scheduler','Configuration','HERO');
job = batch(sched,'test_output');
After termination the job will have written the numbers 1 through 4 to the standard output.
Now, if you want to have a look at that output, you are left with at least two options:
- 1 Use the diary as you would using your local MATLAB installation
- 2 Look up the job related auxiliary files in the jobData folder on your local desktop computer.
Both options will be illustrated below (NOTE: this section was motivated by the user gepp0026).
Using the diary option
Upon submitting a job to the HPC system the CaptureDiary option is set to true by default. If not, you might want to enable it by changing the job description to:
job = batch(sched,'test_output', 'CaptureDiary',true);
Then, after the job has terminated, simply type
diary(job,'myDiary');
to save the standard output within a diary-file called myDiary. Note that the diary is created in the folder you currently reside on with your local MATLAB session. Here, the content of the diary file myDiary reads
Warning: Unable to change to requested folder:
/home/compphys/alxo9476/MATLAB/R2011b/example/myExamples_matlab/MATLAB_EX_testOutput.
Current folder is: /user/fk5/ifp/agcompphys/alxo9476.
Reason: Cannot CD to
/home/compphys/alxo9476/MATLAB/R2011b/example/myExamples_matlab/MATLAB_EX_testOutput
(Name is nonexistent or not a directory).
> In <a href="matlab: > opentoline('/cm/shared/apps/matlab/r2011b/toolbox/distcomp/cluster/+parallel/+internal/+cluster/executeScript.m',12,1)">executeScript > at 12</a>
In <a href="matlab: opentoline('/cm/shared/apps/matlab/r2011b/toolbox/distcomp/private/dctEvaluateFunction.m',21,1)">distcomp/private/dctEvaluateFunction>iEvaluateWithNoErrors at 21</a>
In <a href="matlab: opentoline('/cm/shared/apps/matlab/r2011b/toolbox/distcomp/private/dctEvaluateFunction.m',7,1)">distcomp/private/dctEvaluateFunction at 7</a>
In <a href="matlab: opentoline('/cm/shared/apps/matlab/r2011b/toolbox/distcomp/private/dctEvaluateTask.m',273,1)">distcomp/private/dctEvaluateTask>iEvaluateTask/nEvaluateTask at 273</a>
In <a href="matlab: opentoline('/cm/shared/apps/matlab/r2011b/toolbox/distcomp/private/dctEvaluateTask.m',134,1)">distcomp/private/dctEvaluateTask>iEvaluateTask at 134</a>
In <a href="matlab: opentoline('/cm/shared/apps/matlab/r2011b/toolbox/distcomp/private/dctEvaluateTask.m',58,1)">distcomp/private/dctEvaluateTask at 58</a>
In <a href="matlab: opentoline('/cm/shared/apps/matlab/r2011b/toolbox/distcomp/distcomp_evaluate_filetask.m',161,1)">distcomp_evaluate_filetask>iDoTask at 161</a>
In <a href="matlab: opentoline('/cm/shared/apps/matlab/r2011b/toolbox/distcomp/distcomp_evaluate_filetask.m',64,1)">distcomp_evaluate_filetask at 64</a>
n =
1
n =
2
n =
3
n =
4
The first part is actually a warning message that stems from the client and host being physically separated (i.e. not on the same file system) and not harmful. You can ignore it without worries. The second part lists the command window output of the small application, i.e. the state of the variable n throughout the different iterations.
Looking up the auxiliary job information files
As an alternative to the above option, you might simply look up the job related auxiliary files in your jobData folder on your local desktop computer. Now, just for arguments, say the small job had the jobname Job136. Then, after the job has terminated successfully, there should be a folder called Job136 in your jobData folder. In that particular, job related folder jobData/Job136 you can find a wealth of job related auxiliary files. I don't want to go through them in detail, but here is a list to give you an idea what you should expect to find for a job that employs a single worker:
distributedJobWrapper.sh matlab_metadata.mat Task1.common.mat Task1.in.mat Task1.java.log Task1.jobout.mat Task1.log Task1.out.mat Task1.state.mat tp58a7f63d_5808_49eb_9f71_e4f663be1a3b
Here, in order to access the command window output of the application, the file Task1.out.mat is of importance. Now, to list the command window output you have to do the following: From within your current MATLAB session, navigate to the above folder and open the file Task1.out.mat by typing
load Task1.out.mat
in your MATLAB command window. You can then list the variables in the current workspace (with there respective size and data types) via the command
whos
which, for the above application, yields (if you open up a new MATLAB session for this puropose)
Name Size Bytes Class Attributes argsout 1x1 192 cell commandwindowoutput 1x1759 3518 char erroridentifier 0x0 0 char errormessage 0x0 0 char errorstruct 0x0 56 MException finishtime 1x29 58 char
You might then simply type
commandwindowoutput
to obtain
commandwindowoutput =
Warning: Unable to change to requested folder:
/home/compphys/alxo9476/MATLAB/R2011b/example/myExamples_matlab/MATLAB_EX_testOutput.
Current folder is: /user/fk5/ifp/agcompphys/alxo9476.
Reason: Cannot CD to
/home/compphys/alxo9476/MATLAB/R2011b/example/myExamples_matlab/MATLAB_EX_testOutput
(Name is nonexistent or not a directory).
> In executeScript at 12
In distcomp/private/dctEvaluateFunction>iEvaluateWithNoErrors at 21
In distcomp/private/dctEvaluateFunction at 7
In distcomp/private/dctEvaluateTask>iEvaluateTask/nEvaluateTask at 273
In distcomp/private/dctEvaluateTask>iEvaluateTask at 134
In distcomp/private/dctEvaluateTask at 58
In distcomp_evaluate_filetask>iDoTask at 161
In distcomp_evaluate_filetask at 64
n =
1
n =
2
n =
3
n =
4
Again, the first part is a warning message that stems from the client and host being physically separated (i.e. not on the same file system) and not harmful. You can ignore it without worries. The second part lists the command window output of the small application, i.e. the state of the variable n throughout the different iterations.
Parallelization using parfor loops
Probably the most simple way to parallelize already written code is by using parfor loops. As its name suggests, the command parfor indicates a for loop which is executed in parallel. As soon as your MATLAB session encounters a parfor loop, the range of the iteration variable is subdivided into a specified number of blocks which are in turn processed by different workers.
Note that parfor loops require the iterations to be independent of each other since it is in general not possible to ensure that the iterations finish in a particular order.
Basic example: Vector-Matrix multiplication
To facilitate intuition, consider a vector-matrix multiplication, for given -vector and -matrix , as one would do it by hand: . Or, in terms of a (arguably non-optimal) sequential MATLAB snippet:
for i=1:N
tmp=0.;
for j=1:N
tmp=tmp+A(i,j)*b(j);
end
c(i)=tmp;
end
As evident from that code snipped, the different entries in the solution vector do not depend on each other. In principle, each entry of the solution vector might be computed on a different worker, even at the same time, without affecting the results. Hence, this piece of code might be amended by using a parfor loops instead. A complete example using parfor reads:
N=2048;
b=rand(N,1);
A=rand(N,N);
parfor i=1:N
tmp=0.;
for j=1:N
tmp=tmp+A(i,j)*b(j);
end
c(i)=tmp;
end
In order to submit this example to the HPC system, you need to first save the example as .m-file, e.g. myExample_parfor.m, and then submit it via
sched=findResources('scheduler','Configuration','HERO');
job=batch(sched,'myExample_parfor','matlabpool',8);
Variable slicing
MATLAB has a particular classification scheme for variables within parfor loops (see [1]). A variable might, e.g. be a loop variable, as the variable i in the above example, a temporary variable, as the variable tmp above, or a sliced variable (note that there are more possible classifications). A sliced variable refers to an array whose segments are operated on by different iterations of the loop. Just like for the entries of the output array c. Note that the same holds also for the rows of the input Matrix A: to compute the entry c(i) only the row A(i,:) is needed. In this sense, c(i) is a sliced output variable, while A(i,:) might be used as a sliced input variable. A sliced version of the above example reads:
N=2048; b=rand(N,1); A=rand(N,N); parfor i=1:N c(i)=A(i,:)*b(:); end
Note that parfor loops start by first distributing data to the allocate workers. By using sliced variables only the data which is really needed by a particular worker is sent to it. Hence, the use of sliced variables might result in less overall memory and/or time consumption.
Examples where parfor loops do not apply
Recurrence relations, i.e. equations that define a sequence recursively are, by their very nature, not parallelizable.
Further considerations
In principle, by encountering a parlor loop there is some overhead associated to the startup of parallel processes and to the distribution of data to the workers. Hence, it might not be a good advice to simply replace each loop by a parfor loop. E.g. for the intuitive, yet trivial vector-matrix multiplication example the use of a parlor loop might not pay off, since MATLAB offers efficient implementations for that purpose. Often it pays off to first search for the function that is responsible for the majority of execution time and to parallelize that.
Other Topics
MDCS working mode
On the local HPC facilities we use MATLAB for a non-interactive processing of jobs, i.e. in batch mode, only. The scheduler (we use SGE) offers the dedicated mdcs parallel environment which is used to run a specified number of workers as applications. These applications are started to process user-supplied tasks, and they stop as soon as the tasks are completed. More information on this working mode might be found here.
The parallel environment memory issue
Note that for practically any job you should not request less than the memory specified by default. The reason is related to some memory overheads that accumulate for the master process of your job in case the job gets split across several execution hosts. Consider the following example:
I submitted the above random walk example using the mySubmitScript_v2.m job-submission script. I submitted the job from my local desktop PC, and specified an overall amount of 12 workers to work on the job. The job itself can be considered as a low-memory job, it does not require much memory. Briefly after having submitted the job it is properly displayed via qstat (on HERO):
job-ID prior name user state submit/start at queue slots ja-task-ID --------------------------------------------------------------------------------------------------------- 761093 0.50691 Job131 alxo9476 qw 07/10/2013 12:06:48 12
Unfortunately the job ran and finished successfully while I was off desk. To obtain further information about the competed job I use the qacct tool. Here, for the above job the query qacct -j 761093 yields the output
============================================================== qname mpc_std_shrt.q hostname mpcs001.mpinet.cluster group ifp owner alxo9476 project NONE department defaultdepartment jobname Job131 jobnumber 761093 taskid undefined account sge priority 0 qsub_time Wed Jul 10 12:06:48 2013 start_time Wed Jul 10 13:33:04 2013 end_time Wed Jul 10 13:34:32 2013 granted_pe mdcs slots 12 failed 0 exit_status 0 ru_wallclock 88 ru_utime 94.990 ru_stime 15.927 ru_maxrss 1365624 ru_ixrss 0 ru_ismrss 0 ru_idrss 0 ru_isrss 0 ru_minflt 928976 ru_majflt 11492 ru_nswap 0 ru_inblock 0 ru_oublock 0 ru_msgsnd 0 ru_msgrcv 0 ru_nsignals 0 ru_nvcsw 207366 ru_nivcsw 47240 cpu 110.916 mem 0.003 io 0.003 iow 0.000 maxvmem 818.414M arid undefined
Note that the maximum vmem size of the job turns out to be about 800M. That seems a bit far fetched for such a slim job that uses a negligible amount of memory by itself. So, where does this astronomically high memory consumption (given the low memory job) come from?
First, lets have a look on which execution hosts the job ran. From the above output we already know that the master task ran on host mpcs001, but how many hosts where involved in the process? Albeit there was no chance to use the qstat -g t query to see on which nodes the individual workers where started (since I was off desk during the running time of the program), its possible to get a list of the execution hosts chosen by the scheduler by having a look at the Job131.mpiexec.log file in the respective job folder jobData/Job131/. The very first line of that file contains a list of the execution hosts determined by the scheduler. Broken down in order to facilitate readability, the line reads in my case:
Starting SMPD on mpcs001.mpinet.cluster mpcs005.mpinet.cluster mpcs012.mpinet.cluster mpcs045.mpinet.cluster mpcs056.mpinet.cluster mpcs075.mpinet.cluster mpcs100.mpinet.cluster
So, apparently, the job was split across 7 execution hosts. Effectively, the master task has to maintain a ssh connection to each of the hosts on which the slave tasks are about to run and on which further process have to be started. For each additional host this can easily cost up to 100M-150M that all accumulate for the master task. Hence for 6 additional hosts, the master task has to carry up to 600M-900M. Note that these do not include the computational cost for the actual job, yet. Further, note that the memory overhead accumulates for the master task, only, the slave tasks need less memory.
In any case, if a job involves many workers it is very likely that the job gets split across many execution hosts. Ultimately, this is due to the fill-up allocation rule followed by the scheduler. As illustrated above, in such a case, the master task has to bear up some memory overhead that will result in your job running out of memory and getting killed, if not accounted for.
Permision denied upon attempted passphraseless login on remote host
Consider the following situation: you submit a Job, employing an overall number of three workers, from your local client to the remote HPC system and SGE places the job in an appropriate queue (following your resource specifications). Just for arguments: say, the job got the name Job118. After a while the job changes from its state qw to r and to dr soon after, so that the job is not executed properly.
In your /MATLAB/R2011b/jobData folder on the HPC system (during the runtime of the job), or on your local client (if the job has already finished and the respective files are transfered to your client) there should be a folder called Job118, wherein a log-file can be found. For the job Job118, this log-file is called Job118.mpiexec.log and it might contain some information on why the job is not executed properly. Now, say the content of Job118.mpiexec.log reads something similar to
Starting SMPD on mpcs005.mpinet.cluster mpcs103.mpinet.cluster mpcs111.mpinet.cluster ... /cm/shared/apps/sge/6.2u5p2/mpi/mdcs/start_mdcs -n mpcs005.mpinet.cluster "/cm/shared/apps/matlab/r2011b/bin/mw_smpd" 25327 /cm/shared/apps/sge/6.2u5p2/mpi/mdcs/start_mdcs -n mpcs103.mpinet.cluster "/cm/shared/apps/matlab/r2011b/bin/mw_smpd" 25327 /cm/shared/apps/sge/6.2u5p2/mpi/mdcs/start_mdcs -n mpcs111.mpinet.cluster "/cm/shared/apps/matlab/r2011b/bin/mw_smpd" 25327 Permission denied, please try again. Permission denied, please try again. Check for smpd daemons (1 of 10) Permission denied (publickey,gssapi-with-mic,password). Warning: Permanently added 'mpcs005.mpinet.cluster,10.142.3.5' (RSA) to the list of known hosts. Warning: Permanently added 'mpcs111.mpinet.cluster,10.142.3.111' (RSA) to the list of known hosts. Missing smpd on mpcs005.mpinet.cluster Missing smpd on mpcs103.mpinet.cluster Missing smpd on mpcs111.mpinet.cluster Permission denied, please try again. Permission denied, please try again. Permission denied, please try again. Permission denied, please try again. Permission denied (publickey,gssapi-with-mic,password). Permission denied (publickey,gssapi-with-mic,password). Check for smpd daemons (2 of 10) Missing smpd on mpcs005.mpinet.cluster Missing smpd on mpcs103.mpinet.cluster Missing smpd on mpcs111.mpinet.cluster
This might indicate that the passphraseless login to the execution nodes, here the nodes mpcs005.mpinet.cluster mpcs103.mpinet.cluster mpcs111.mpinet.cluster on which the workers should be started, has failed.
Albeit there are many reasons that might cause such a behavior, one that seems likely is the following: you probably changed the rights of your home directory in a way that also the group and/or others have the right to write to your home directory. To make this as explicit as possible, I change my rights accordingly to give an example: so, in my homedirectory on HERO I type
ls -lah ./
to find
drwxrwxrwx 14 alxo9476 ifp 937 Jul 2 12:19 .
considering the ten leftmost letters this is bad. The first d tells that we are looking at a directory. The following three letters rwx tell that the owner of the directory has the rights to read, write, and execute. The following three letters signify that the group has the right to read, write, and execute and the last three letters indicate that all others have the right to read, write, and execute files. Note that for a successful passphraseless login on a remote host, it must be made sure that only the owner of the remote users home directory (which in this case are you, yourself!), has the right to write.
As a remedy try the following: To resore the usual rights in my homedirectory on HERO I type
chmod 755 ./
and now find
drwxr-xr-x 14 alxo9476 ifp 937 Jul 2 12:19 .
which shows that the writing rights for the unix group I belong to (in my case ifP) and all others are removed. Accordingly, a passphraseless login to the hosts specified in the Hostfile should now be possible. I.e., after resubmission of the job it should run properly.
Further troubleshooting
Say you submitted a job, here referred to as Job1, several times already but it does not finish successfully. Now, if you have no idea how to troubleshoot further and would like to inform the coordinator of scientific computing about such problems in order to ask for help, it would be beneficial and would help in the process of finding the problem if you could provide the following:
- Job1.mpiexec.log-file: If you configured your profile according to the mdcs wiki page you should find a folder called Job1 in the jobData folder, local to your desktop computer. Within that folder there should be a log-file, in this case called Job1.mpiexec.log, e.g., containing information on the nodes that were allocated by the scheduler, the startup/shutdown of smpd daemons on the execution host and the launch of the actual task function you submitted, etc.
- Job-Id: If you happened to have a look at the status of the job via the command qstat you also might have recognized the job-id of the job. If you could also provide the job-id, that would even help further.